
Examples for count outcome
Vignette 1 of 4
July 03, 2025
Source:vignettes/Count-examples.Rmd
Count-examples.Rmdprecmed: Precision Medicine in R
A doubly robust precision medicine approach to estimate and validate conditional average treatment effects
Applying precmed to count outcome data
Example dataset
We consider a simulated dataset that is based on real-world claims
data from patients with multiple sclerosis. The dataset
countExample includes 4,000 patients with information on
the following 9 variables.
| level | Overall | drug0 | drug1 | |
|---|---|---|---|---|
| Number of patients | 4000 | 998 | 3002 | |
| Age (mean (SD)) | 46.2 (9.8) | 46.1 (10.1) | 46.3 (9.7) | |
| Gender (%) | male | 987 (24.7) | 247 (24.7) | 740 (24.7) |
| female | 3013 (75.3) | 751 (75.3) | 2262 (75.3) | |
| Previous treatment (%) | drugA | 1787 (44.7) | 457 (45.8) | 1330 (44.3) |
| drugB | 435 (10.9) | 110 (11.0) | 325 (10.8) | |
| drugC | 1778 (44.5) | 431 (43.2) | 1347 (44.9) | |
| Previous medical cost ($) (mean (SD)) | 13823.8 (20172.3) | 14181.0 (21591.3) | 13705.1 (19680.3) | |
| Previous number of symptoms (%) | 0 | 332 ( 8.3) | 73 ( 7.3) | 259 ( 8.6) |
| 1 | 2669 (66.7) | 662 (66.3) | 2007 (66.9) | |
| >=2 | 999 (25.0) | 263 (26.4) | 736 (24.5) | |
| Previous number of relapses (%) | 0 | 2603 (65.1) | 646 (64.7) | 1957 (65.2) |
| 1 | 1114 (27.9) | 280 (28.1) | 834 (27.8) | |
| 2 | 245 ( 6.1) | 59 ( 5.9) | 186 ( 6.2) | |
| 3 | 32 ( 0.8) | 12 ( 1.2) | 20 ( 0.7) | |
| 4 | 4 ( 0.1) | 1 ( 0.1) | 3 ( 0.1) | |
| 5 | 2 ( 0.0) | 0 ( 0.0) | 2 ( 0.1) | |
| Number of relapses during follow-up (%) | 0 | 3086 (77.1) | 763 (76.5) | 2323 (77.4) |
| 1 | 640 (16.0) | 170 (17.0) | 470 (15.7) | |
| 2 | 173 ( 4.3) | 49 ( 4.9) | 124 ( 4.1) | |
| 3 | 54 ( 1.4) | 9 ( 0.9) | 45 ( 1.5) | |
| 4 | 30 ( 0.8) | 5 ( 0.5) | 25 ( 0.8) | |
| 5 | 8 ( 0.2) | 1 ( 0.1) | 7 ( 0.2) | |
| 6 | 5 ( 0.1) | 1 ( 0.1) | 4 ( 0.1) | |
| 7 | 2 ( 0.0) | 0 ( 0.0) | 2 ( 0.1) | |
| 8 | 2 ( 0.0) | 0 ( 0.0) | 2 ( 0.1) | |
| Length of follow-up (years) (mean (SD)) | 1.2 (1.2) | 1.2 (1.3) | 1.2 (1.2) |
Most variables are baseline patient characteristics like age, gender, and previous treatment.
We will use
yas the count outcome variable, which is the number of relapses during follow-up.We will use
yearsas the offset variable, which is number of years during follow-up.We will use
trtas the treatment variable, which has 2 drugs (drug0 and drug1).
To avoid multicollinearity issues, the continuous variable
age was centered at 48 years old and the medical costs in
the year prior to treatment initiation previous_cost was
centered at 13,824 USD and scaled with standard deviation 20,172
USD.
The final dataset countExample looked like this:
#> 'data.frame': 4000 obs. of 9 variables:
#> $ age : num [1:4000, 1] -21.22 -2.22 -10.22 6.78 4.78 ...
#> ..- attr(*, "scaled:center")= num 46.2
#> $ female : int 0 1 1 0 1 0 1 0 1 1 ...
#> $ previous_treatment : Factor w/ 3 levels "drugA","drugB",..: 1 1 1 1 3 3 3 3 3 1 ...
#> $ previous_cost : num [1:4000, 1] 0.451 -0.194 -0.534 -0.337 -0.271 ...
#> ..- attr(*, "scaled:center")= num 13824
#> ..- attr(*, "scaled:scale")= num 20172
#> $ previous_number_symptoms: Factor w/ 3 levels "0","1",">=2": 2 2 2 2 2 2 2 3 2 1 ...
#> $ previous_number_relapses: int 0 0 1 1 0 0 0 0 1 1 ...
#> $ trt : Factor w/ 2 levels "drug0","drug1": 2 1 1 2 2 1 2 2 2 2 ...
#> $ y : int 0 0 0 0 1 0 0 0 0 0 ...
#> $ years : num 0.2847 0.6105 2.653 0.0383 2.2697 ...Estimation of the ATE with atefit()
First, one might be interested in the effect of the drug (trt) on the number of relapses during follow-up (y). Lets test this using simple regression.
output_lm <- lm(y ~ trt, countExample)
output_lm
#>
#> Call:
#> lm(formula = y ~ trt, data = countExample)
#>
#> Coefficients:
#> (Intercept) trtdrug1
#> 0.32665 0.02045We see from the simple linear model that drug 1 scores 0.02 points higher compared to drug 0. This indicates that drug 0 is superior to drug 1 because lower outcomes are preferred (number of relapses).
Now we want to estimate the Average Treatment Effect (ATE) and
correct for several covariates like age and previous treatment because
they might influence the relation between treatment and outcome. The
atefit() function allows estimating the ATE in terms of
rate ratio. The rate ratio estimator is doubly robust, meaning that the
estimator is consistent if the propensity score (PS) model (argument
ps.model) or the outcome model (argument
cate.model) or both are correctly specified. The function
also provides standard error, confidence intervals, and p-values based
on bootstrap.
The mandatory arguments for atefit() are:
response: “count” in this example because we use have data with a count outcome (e.g., number of relapses))data: the name of the data set. (countExample)cate.model: A formula describing the outcome model to be fitted. The outcome must appear on the left-hand side. In the example, we choose to specify to outcome model as a linear combination of the following covariates: age, sex, previous treatment, medical costs in the year prior to treatment initiation, and number of relapses in the year prior to treatment initiation. Non-linear or interaction terms could also be included. The outcome model has the offsetlog(years)to account for the varying exposure times across patients. Note that the treatment variable is not supplied incate.modelsince this is an outcome model.
-
ps.model: A formula describing the propensity score (PS) model to be fitted. The treatment must appear on the left-hand side and the covariates (age and previous treatment in this example) on the right-hand side. The variabletrtmust be supplied as a numeric variable taking only 2 values, 1 for active treatment and 0 for control or comparator. If it is not the case, the function will stop with error iftrttakes more than 2 distinct values or will automatically transformtrtinto a numeric variable. In this example,trt(a factor variable taking values “drug0” and “drug1”) was transformed and a warning message was left to the user (see output below):Variable trt was recoded to 0/1 with drug0->0 and drug1->1. If the data are from a RCT, it suffices to specifyps.model= trt ~ 1. Note that the PS model is only used in the estimation of the 2 doubly robust methods (two and contrast regressions).
Let’s calculate the ATE for the effect of treatment on y and build an
outcome model that contains all variables and a PS model with
age and previous_treatment.
output_atefit <- atefit(response = "count",
data = countExample,
cate.model = y ~ age + female + previous_treatment + previous_cost + previous_number_relapses + offset(log(years)),
ps.model = trt ~ age + previous_treatment,
n.boot = 50,
seed = 999,
verbose = 0)
#> Warning in data.preproc(fun = "drinf", cate.model = cate.model, ps.model = ps.model, : Variable trt was recoded to 0/1 with drug0->0 and drug1->1.When verbose = 1, the function outputs a progress bar in
the console.
output_atefit
#> Average treatment effect:
#>
#> estimate SE CI.lower CI.upper pvalue
#> log.rate.ratio 0.06311746 0.1624482 -0.2552752 0.3815101 0.6976173
#>
#> Estimated event rates:
#>
#> estimate
#> rate1 0.2860635
#> rate0 0.2685660
#> Warning in print.atefit(x): Variable trt was recoded to 0/1 with drug0->0 and drug1->1.The output of atefit() shows the point estimate,
standard error (“SE”), lower (“CI.lower”) and upper (“CI.upper”) bound
of the 95% confidence interval, and the p-value (“pvalue”) for the log
rate ratio ($log.rate.ratio) as well as the point estimate
for the rate in the 2 treatment groups ($rate0 and
$rate1). For example, the log rate ratio of 0.06 and the
95% confidence interval of (-0.26, 0.38) are displayed in the output.
The user can retrieve the rate ratio to facilitate the
interpretation:
rate.ratio <- exp(output_atefit$log.rate.ratio$estimate)
rate.ratio
#> [1] 1.065152
CI.rate.ratio <- exp(output_atefit$log.rate.ratio$estimate + c(-1, 1) * qnorm(0.975) * sqrt(output_atefit$log.rate.ratio$SE))
CI.rate.ratio
#> [1] 0.4834326 2.3468602The rate ratio of 1.07 along with the 95% confidence interval of
(0.48, 2.35) suggest that drug 0 is superior to drug 1 because the ratio
is greater than 1 and lower outcomes are preferred, but that this
superiority is not statistically significant given the p-value of 0.7 in
the output ($log.rate.ratio$pvalue).
Using plot(output_atefit), a histograms is generated of
the point estimates across the n.boot bootstrap iterations
for the log rate ratio. A red vertical line is added to each histogram
with the mean of the bootstrap estimates.
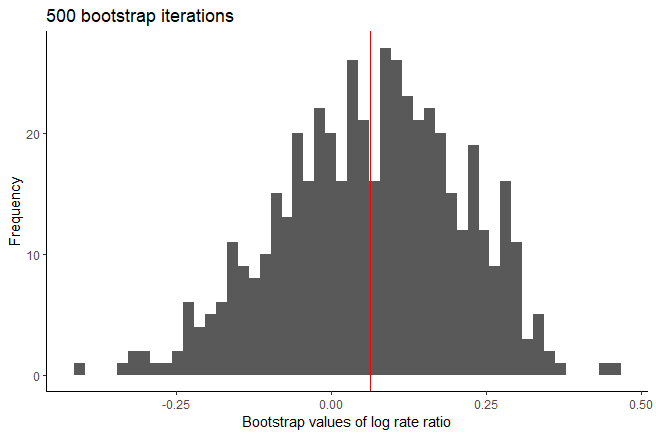
Histograms of bootstrap log rate ratio estimators after 500 bootstrap iterations.
Estimation of the CATE score with catefit()
Now we have calculated the ATE we know the effect of both treatments
on average, but this effect might not be the same for all patients.
Thus, it might be worthwhile to check if there are subgroups that
respond differently to the treatments. We can calculate the Conditional
Average Treatment Effect (CATE) score which calculates the ATE for
different subgroups in the data. If no internal validation is needed (we
get back to that later), we can use the catefit() function
to a model directly to the entire data set to estimate the CATE score.
For the catefit() function we have to define the
score.method. This arguments specifies the precision
medicine (PM) method to be used to calculate the CATE scores. There are
a total of 5 scoring methods implemented:
poissonfits a Poisson model separately by treatment group.boostinguses gradient boosted regression models (GBM) separately by treatment group.twoRegimplements the doubly robust two regressions estimator in (Yadlowsky et al. 2020).contrastRegimplements the doubly robust contrast regression estimator from (Yadlowsky et al. 2020).negBinfits negative binomial regressions by treatment group. This method is recommended if there is overdispersion in the data.
When we have selected a PS method that fits the data (or multiple
methods), we can run the catefit() function with the same
variables as the atefit() function. The mandatory arguments
are: response, data,
score.method, cate.model, and
ps.model. The user can also specify the non-mandatory
arguments to fit with the data and problem at hand. Please see the Function
description section for details.
If you run into errors or warnings with your data, it might be helpful to go over the descriptions to see if you need to alter the default values. In this toy example, we keep the default values of the remaining arguments.
t0 <- Sys.time()
output_catefit <- catefit(response = "count",
data = countExample,
score.method = c("poisson", "boosting", "twoReg", "contrastReg", "negBin"),
cate.model = y ~ age + female + previous_treatment + previous_cost + previous_number_relapses + offset(log(years)),
ps.model = trt ~ age + previous_treatment,
initial.predictor.method = "poisson",
higher.y = FALSE,
seed = 999)
#> Warning in data.preproc(fun = "catefit", cate.model = cate.model, ps.model = ps.model, : Variable trt was recoded to 0/1 with drug0->0 and drug1->1.
t1 <- Sys.time()
t1 - t0
#> Time difference of 13.48409 secsEach method specified in score.method has the following
sets of results in catefit():
-
scorecontains the log-transformed estimated CATE scores for each subject. CATE score is a linear combination of the variables specified in thecate.modelargument. Same as the outcome, lower CATE scores are more desirable ifhigher.y= FALSE and vice versa. Each subject has one CATE score so the length of this output is 4,000 for our toy example. Below we show the CATE scores estimated with contrast regression for the first 6 subjects in the data.
length(output_catefit$score.contrastReg)
#> [1] 4000
head(output_catefit$score.contrastReg)
#> [1] 0.1497829 0.2624196 0.5864289 -0.8024851 -0.1907681 -0.4049082-
coefficientscontains the estimated coefficients of the CATE score for each scoring method. It is a data frame of the covariates (including intercept) as rows and scoring methods as columns. In our toy example, there are 5 covariates in thecate.model(including a categorical variable with 3 distinct factors) so there are 7 rows of estimated coefficients within each column. Since contrast regression is one of the scoring methods specified in the example, we see that contrast regression has an additional column with the standard errors of the estimated coefficients. Boosting does not estimate coefficients (it directly predicts the score) so there is no coefficient result for this method.
output_catefit$coefficients
#> poisson twoReg contrastReg SE_contrastReg
#> (Intercept) -0.58185218 -0.57230335 -0.59733948 0.37239529
#> age -0.01117701 -0.04006858 -0.03579484 0.01382861
#> female 0.58829733 0.73791026 0.77499335 0.36588639
#> previous_treatmentdrugB 0.73724361 0.64116126 0.74877593 0.35907273
#> previous_treatmentdrugC 0.15545985 -0.25808471 -0.20473293 0.26470389
#> previous_cost -0.13035642 -0.08457179 -0.02750876 0.15712507
#> previous_number_relapses -0.05339112 0.06117043 0.02831604 0.16882933
#> negBin
#> (Intercept) -0.59232437
#> age -0.01410487
#> female 0.59602219
#> previous_treatmentdrugB 0.76032497
#> previous_treatmentdrugC 0.13262969
#> previous_cost -0.13952096
#> previous_number_relapses -0.05034136We can define the estimated CATE scores for contrast regression like shown below. The user can use this information to study the influence of each covariate.
-
atecontains estimated ATEs in each nested subgroup of high responders to drug 1 defined byprop.cutoff. The subgroups are defined based on the estimated CATE scores with the specified scoring method. In this example, we show the estimated ATEs of subgroups identified by CATE scores of contrast regression. For example, the estimated ATE for the subgroup of subjects constructed based on the 50% (“prop0.5”) lowest CATE scores estimated from contrast regression is 0.62.
output_catefit$ate.contrastReg
#> prop0.5 prop0.6 prop0.7 prop0.8 prop0.9 prop1
#> 0.6191191 0.6630810 0.7910504 0.8432274 0.9372587 1.0640579You are encouraged to summarize and visualize the outputs in
whichever way that fits a particular situation outside the package’s
functions. For example, it is possible to plot the densities of all CATE
scores with ggplot(). There are some subjects with
extremely low CATE scores but most of the samples fall between -1 and 1
with triple modes at around -0.5, 0, and 0.8.
dataplot <- data.frame(score = factor(rep(c("Boosting", "Naive Poisson", "Two regressions", "Contrast regression", "Negative Binomial"), each = length(output_catefit$score.boosting))),
value = c(output_catefit$score.boosting, output_catefit$score.poisson, output_catefit$score.twoReg, output_catefit$score.contrastReg, output_catefit$score.negBin))
dataplot %>%
ggplot(aes(x = value, fill = score)) +
geom_density(alpha = 0.5) +
theme_classic() +
labs(x = "Estimated CATE score", y = "Density", fill = "Method")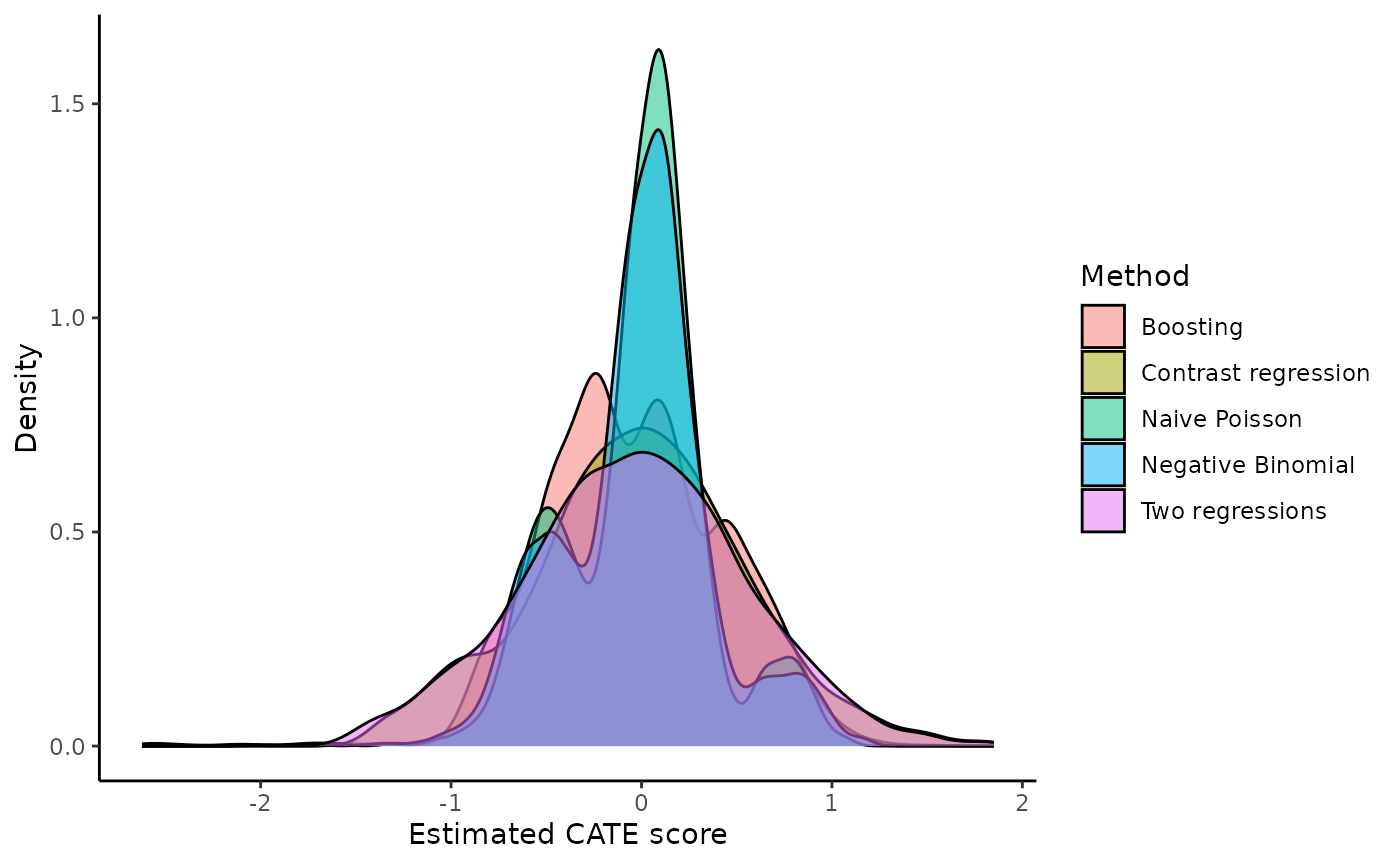
Internal validation via catecv()
The catecv() function provides the same estimation as
the catefit() but via cross-validation (CV). With the
catecv() function internal CV is applied to reduce optimism
in choosing the CATE estimation method that captures the most treatment
effect heterogeneity. The CV is applied by repeating the following steps
cv.n times:
Split the data into a training and validation set according to
train.propargument. The training and validation sets must be balanced with respect to covariate distributions and doubly robust rate ratio estimates (seeerror.maxargument).Estimate the CATE score in the training set with the specified scoring method.
Predict the CATE score in the validation set using the scoring model fitted from the training set.
-
Build nested subgroups of treatment responders in the training and validation sets, separately, and estimate the ATE within each nested subgroup. For each element i of
prop.cutoffargument (e.g.,prop.cutoff[i] = 0.6), take the following steps:Identify high responders as observations with the 60% (i.e.,
prop.cutoff[i]x100%) highest (ifhigher.y = TRUE) or lowest (ifhigher.y = FALSE) estimated CATE scores.Estimate the ATE in the subgroup of high responders using a doubly robust estimator.
Conversely, identify low responders as observations with the 40% (i.e.,
1 - prop.cutoff[i]x100%) lowest (ifhigher.y = TRUE) or highest (ifhigher.y = FALSE) estimated CATE scores.Estimate the ATE in the subgroup of low responders using a doubly robust estimator.
If
abc = TRUE, calculate the area between the ATE and the series of ATEs in nested subgroups of high responders in the validation set. (for more information about the abc score, see Validation curves and the ABC statistics)Build mutually exclusive subgroups of treatment responders in the training and validation sets, separately, and estimate the ATE within each subgroup. Mutually exclusive subgroups are built by splitting the estimated CATE scores according to prop.multi.
Oke, so now we can use the catecv() function to run
internal validation to compare the different scoring methods. The
mandatory arguments are similar to atefit() and
catefit(): The mandatory arguments are:
response, data, score.method,
cate.model, and ps.model. For this toy example
we selected poisson, contrastReg and
negBin to limit run time.
We also specified the following non-mandatory arguments to fit with
the data and problem at hand: initial.predictor.method,
higher.y, cv.n, seed,
plot.gbmperf and verbose.
initial.predictor.methodspecifies how predictions of the outcome are estimated in two regressions and contrast regression. Flexible models can be used such as GBM (“boosting”) or generalized additive models (“gam”). Both methods are computationally intensive so we choose “poisson” to obtain predictions from a Poisson regression, which reduces the computational time at the expense of stricter parametric assumptions and less flexibility.higher.ywas set to FALSE because lower number of relapses are more desirable in our example. Hence, we are telling the function that subgroups of high responders to drug1 vs drug0 should have lower number of relapses (see section Validation curves and the ABC statistics for illustration). In other situation, higher outcomes may be more favorable, for example, walking more steps in a study on physical activity. It is important for this argument to match with theyoutcome because it will affect how the subgroups are defined by the CATE scores and the performance metrics.We perform 5 CV iterations by specifying
cv.n= 5. Typically, more CV iterations are desirable although associated with longer computational times.We set a random seed
seed= 999 to reproduce the results.We avoid generating the boosting performance plots by specifying
plot.gbmperf= FALSE.When
verbose= 1, progress messages are printed in the R console but errors and warnings are not printed. The current CV iteration is printed, followed by the steps of the CV procedure (splitting the data, training the models, validating the models) and warnings or errors that have occurred during the steps (none in this example). A timestamp and a progress bar are also displayed upon completion of a CV iteration. IfcontrastRegwas selected as one of the methods inscore.method, an additional line of output message will indicate whether the algorithm has converged.
There are many other non-mandatory arguments that
catecv() can accept. Please see the Additional examples vignette for
more examples and the Function
description section for details. If you run into errors or warnings
with your data, it might be helpful to go over the descriptions to see
if you need to alter the default values. In this toy example, we keep
the default values of the remaining arguments.
output_catecv <- catecv(response = "count",
data = countExample,
score.method = c("poisson", "contrastReg", "negBin"),
cate.model = y ~ age + female + previous_treatment + previous_cost + previous_number_relapses + offset(log(years)),
ps.model = trt ~ age + previous_treatment,
initial.predictor.method = "poisson",
higher.y = FALSE,
cv.n = 5,
seed = 999,
plot.gbmperf = FALSE,
verbose = 1)
#> Warning in data.preproc(fun = "crossv", cate.model = cate.model, ps.model = ps.model, : Variable trt was recoded to 0/1 with drug0->0 and drug1->1.
#> | | | 0%
#> cv = 1
#> splitting the data..
#> training..
#> validating..
#> convergence TRUE
#> Thu Jul 3 14:03:41 2025
#> | |============== | 20%
#> cv = 2
#> splitting the data..
#> training..
#> validating..
#> convergence TRUE
#> Thu Jul 3 14:03:45 2025
#> | |============================ | 40%
#> cv = 3
#> splitting the data..
#> training..
#> validating..
#> convergence TRUE
#> Thu Jul 3 14:03:51 2025
#> | |========================================== | 60%
#> cv = 4
#> splitting the data..
#> training..
#> validating..
#> convergence TRUE
#> Thu Jul 3 14:03:57 2025
#> | |======================================================== | 80%
#> cv = 5
#> splitting the data..
#> training..
#> validating..
#> convergence TRUE
#> Thu Jul 3 14:04:04 2025
#> | |======================================================================| 100%
#> Total runtime : 31.13 secsThe output of catecv() is an object of class “precmed”
and here we named it output_catecv. It carries the relevant
information to use in the next step of the workflow which selects the
method (among those specified in the argument score.method)
capturing the highest level of treatment effect heterogeneity. The
output, which is described below, will be used in the functions
abc(), plot() and boxplot().
For each method specified in the argument score.method,
the following 3 groups of outputs are generated: high,
low and group. We use the results from
contrastReg as an example.
1. ATEs in nested subgroups of high responders
This output stores the ATEs - the ratio of annualized relapse rate
between drug1 vs drug0 in this example - in nested subgroups of patients
of high responders to drug 1 in the training
($ate.est.train.high.cv) and validation
($ate.est.valid.high.cv) sets across all CV iterations. For
count outcomes, when higher.y = TRUE, higher CATE scores
correspond to high responders to drug1. When higher.y =
FALSE, lower CATE scores correspond to high responders to drug1. Note
that this is different for survival outcomes. The direction of CATE
scores depends on both higher.y and outcome type.
output_catecv$ate.contrastReg$ate.est.train.high.cv
#> cv1 cv2 cv3 cv4 cv5
#> prop0.5 0.5896053 0.6428854 0.5744973 0.6406698 0.5924975
#> prop0.6 0.6744447 0.7114461 0.6589297 0.6930484 0.7186517
#> prop0.7 0.7383527 0.8360698 0.7806273 0.7779180 0.7539283
#> prop0.8 0.8034239 0.9092085 0.8208183 0.8263584 0.8117300
#> prop0.9 0.9041969 1.0257463 0.9595834 0.9049020 0.9452626
#> prop1 1.0599740 1.0520675 1.0577453 1.0641114 1.0677619
output_catecv$ate.contrastReg$ate.est.valid.high.cv
#> cv1 cv2 cv3 cv4 cv5
#> prop0.5 0.7448829 0.6271739 0.8552290 0.5565145 0.5514821
#> prop0.6 0.7325445 0.7194496 0.8308576 0.5906053 0.6909427
#> prop0.7 0.9166009 0.7932235 0.8468713 0.8076085 0.7944857
#> prop0.8 1.1280003 0.8535919 0.9563156 0.9041068 0.8696296
#> prop0.9 1.1300157 0.8374393 0.9452552 0.9264986 0.9791587
#> prop1 1.0812603 1.0978549 1.0531927 1.0726353 1.0689638The output is a matrix with columns corresponding to the CV
iterations, labeled from 1 to cv.n, and rows corresponding
to nested subgroups. The nested subgroups of patients are defined by the
argument prop.cutoff. Here, we use the default
seq(0.5, 1, length = 6) which defines 6 nested subgroups
with the 50%, 60%, 70%, 80%, 90% and 100% lowest (highest if
higher.y = TRUE) CATE scores estimated by contrast
regression. The rows in the output are labeled to reflect the
user-specified proportions used to build the subgroups.
For example, in the training set and in the first CV iterations (first column labeled “cv1”), the subgroup defined with the 50% lowest CATE scores (first row labeled “prop0.5”) has an estimated RR of 0.59. In contrast, the subgroup defined with all patients (last row labeled “prop1”) has an estimated RR of 1.06.
2. ATEs in nested subgroups of low responders
This output stores the ATEs in nested subgroups of low
responders to drug1 in the training
($ate.est.train.low.cv) and validation
($ate.est.valid.low.cv) sets across all CV iterations. For
count outcomes, when higher.y = TRUE, lower CATE scores
correspond to low responders to drug1. When higher.y =
FALSE, higher CATE scores correspond to low responders to drug1. Again,
this is different for survival outcomes. The direction of CATE scores
depends on both higher.y and outcome type.
output_catecv$ate.contrastReg$ate.est.train.low.cv
#> cv1 cv2 cv3 cv4 cv5
#> prop0.5 1.619590 1.556901 1.657101 1.542445 1.623386
#> prop0.4 1.710009 1.575580 1.727388 1.608509 1.594635
#> prop0.3 1.850595 1.542156 1.629855 1.648721 1.737306
#> prop0.2 1.919854 1.573453 1.961972 1.853781 2.122016
#> prop0.1 2.077797 1.254235 1.522567 2.118723 2.074404The outputs are also matrices with columns corresponding to the CV iterations and rows corresponding to nested subgroups.
The output for the low responders brings additional information to
the user. It gives the ATEs in the complement of each nested subgroup of
high responders. For example, the complement of the subgroup of high
responders defined as patients with the 60% lowest (highest if
higher.y = TRUE) estimated CATE scores is the subgroup low
responders defined as patients with the 40% highest (lowest if
higher.y = TRUE) estimated CATE scores, labeled as
“prop0.4”. In the training set and in the first CV iterations, the
estimated RR is 0.674 in the 60% high responders to drug 1 and 1.71 in
the 40% low responders.
3. ATEs in mutually exclusive subgroups
This output stores the ATEs in mutually exclusive multi-category
subgroups of patients in the training
($ate.est.train.group.cv) and validation
($ate.est.valid.group.cv) sets across all CV
iterations.
output_catecv$ate.contrastReg$ate.est.train.group.cv
#> cv1 cv2 cv3 cv4 cv5
#> prop0.33 1.8635694 1.497846 1.7138028 1.6908566 1.649720
#> prop0.67 1.0699029 1.489476 1.0511935 1.1809557 1.111132
#> prop1 0.4831034 0.470482 0.4764857 0.4816721 0.535003
output_catecv$ate.contrastReg$ate.est.valid.group.cv
#> cv1 cv2 cv3 cv4 cv5
#> prop0.33 1.4220392 2.0022062 1.3647971 1.5853124 1.9901237
#> prop0.67 0.7328152 0.7720613 1.1046447 0.9853931 0.9878052
#> prop1 0.8122051 0.6583514 0.6605675 0.5232875 0.5790997The output is a matrix with columns corresponding to the CV
iterations and rows corresponding to the mutually exclusive subgroups.
The previous 2 outputs only focus on binary subgroups (high or low
responders). Here, the mutually exclusive subgroups can be more than 2
and are defined by the argument prop.multi. We use the
default c(0, 1/3, 2/3, 1) which defines 3 subgroups of
patients with the 33% lowest, 33% middle and 33% highest estimated CATE
scores when higher.y = FALSE (as in this example), or with
the 33% highest, 33% middle and 33% lowest estimated CATE scores when
higher.y = FALSE. Taking the first column as an example,
the first CV iteration calculated 1.864 as the RR for the subgroup with
the 33% lowest estimated CATE scores, 1.07 as the RR for subgroup with
the 33% middle estimated CATE scores, and 0.483 as the RR for subgroup
with the 33% highest estimated CATE scores.
Comparison of methods with abc()
The ABC statistics is calculated by abc() for each
scoring method specified in catecv() and for each of the
cv.n CV iterations using the output object
output_catecv from catecv(). The ABC
corresponds to the area between the curve formed by the ATEs in
subgroups of high responders in the validation set (e.g.,
output_catecv$ate.contrastReg$ate.est.valid.cv for contrast
regression) and the horizontal line representing the ATE in the
validation set. A higher ABC value means that the method captures more
treatment effect heterogeneity. See the Validation curves and the ABC
statistics section for a detailed illustration of the relationship
between higher.y, abc, and the validation
curves.
output_abc <- abc(x = output_catecv)
output_abc
#> cv1 cv2 cv3 cv4 cv5
#> poisson 0.13758995 0.2236276 0.06938081 0.09647510 0.1304204
#> contrastReg 0.07311255 0.1602017 0.06894946 0.14832787 0.1310745
#> negBin 0.14517127 0.2203584 0.08899030 0.09382214 0.1395976The output is a matrix with columns corresponding to the CV
iterations and rows corresponding to the scoring methods specified in
score.method. For example, in CV iteration 1, negative
binomial (“negBin”) has an ABC of 0.145, which is the highest in this CV
iteration, meaning that negative binomial offers the best performance in
the first CV iteration. The user can combine the ABC for each method
across iterations:
average_abc <- apply(output_abc, 1, mean)
average_abc
#> poisson contrastReg negBin
#> 0.1314988 0.1163332 0.1375879In this example, negative binomial also offers the best overall performance because it has the highest average ABC, followed closely by Poisson.
Visualization of the validation curves with plot()
The ATEs of nested subgroups of high responders to drug1 (e.g.,
output_catecv$ate.contrastReg$ate.est.train.high.cv and
output_catecv$ate.contrastReg$ate.est.valid.high.cv for
contrast regression) can be visualized as a side-by-side line plot, with
training results on the left and validation results on the right. The
x-axis is determined by prop.cutoff and the y-axis is the
estimated ATEs averaged over cv.n CV iterations as
specified by cv.i = NULL. The estimated ATE is expressed as
a RR of drug1 versus drug0 for our toy example. By default, the function
retrieves the name of the treatment variable (trt) and the
original labels (drug0 and drug1) to specify a
meaningful y-axis label. Otherwise, it is possible to customize the
y-axis label via the ylab, for example, by using
Rate ratio of drug1 vs drug0 in each subgroup.
Steeper slopes indicate more treatment effect heterogeneity between
drug1 and drug0. Because higher.y = FALSE in this example,
the slopes should be increasing from left (prop.cutoff =
0.5) to right (prop.cutoff = 1) if treatment effect
heterogeneity is present. The method that has the steepest slope in the
validation results would be selected because it captures the most
treatment effect heterogeneity while generalizing well to unseen
data.
plot(x = output_catecv)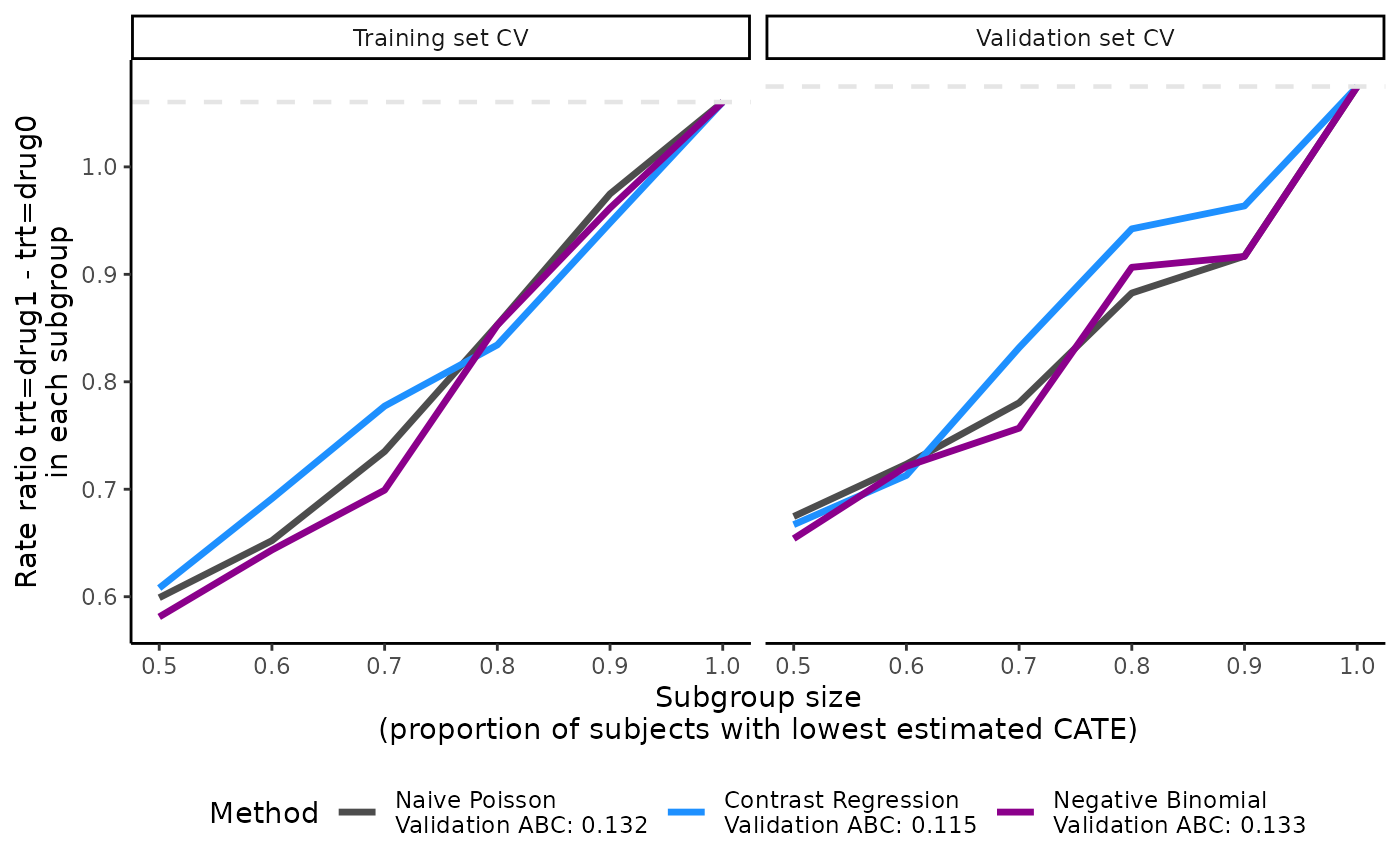
For this toy example, the methods are performing well in the training
data as per the steep, increasing slopes on the left plot. Moreover, all
methods generalize well to the validation data, as indicated by the
monotonous increasing curves in the validation data (right plot). The
dashed gray line is the ATE in the entire data set, which is why all
lines merge to this reference line when subgroup size is 100% of the
data (prop.cutoff = 1). For more explanation on the
validation curves, see the Function
description section.
The plot’s legend includes the ABC statistics in the validation set.
The user can choose to mute the ABC annotations by specifying
show.abc = FALSE.
plot(x = output_catecv,
show.abc = FALSE,
ylab = c("Rate ratio of drug1 vs drug0 in each subgroup"))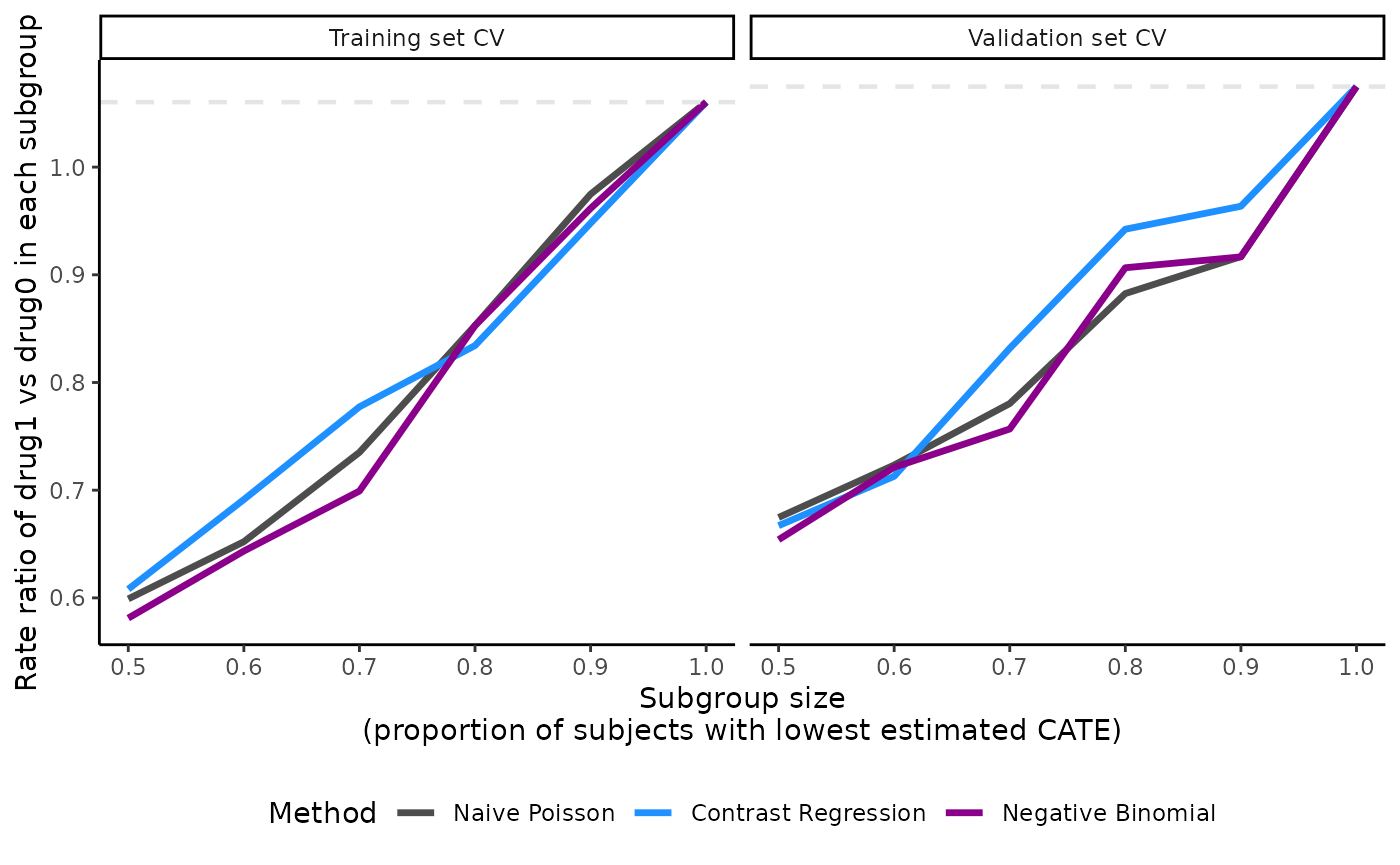
The user can choose to plot the validation curves of only one CV
iteration instead of the average of all CV iterations. In the following
example, we plot the validation curves of the second CV iteration by
specifying cv.i = 2 and in grayscale by specifying
grayscale = TRUE.
plot(x = output_catecv,
cv.i = 2,
grayscale = TRUE,
ylab = c("Rate ratio of drug1 vs drug0 in each subgroup"))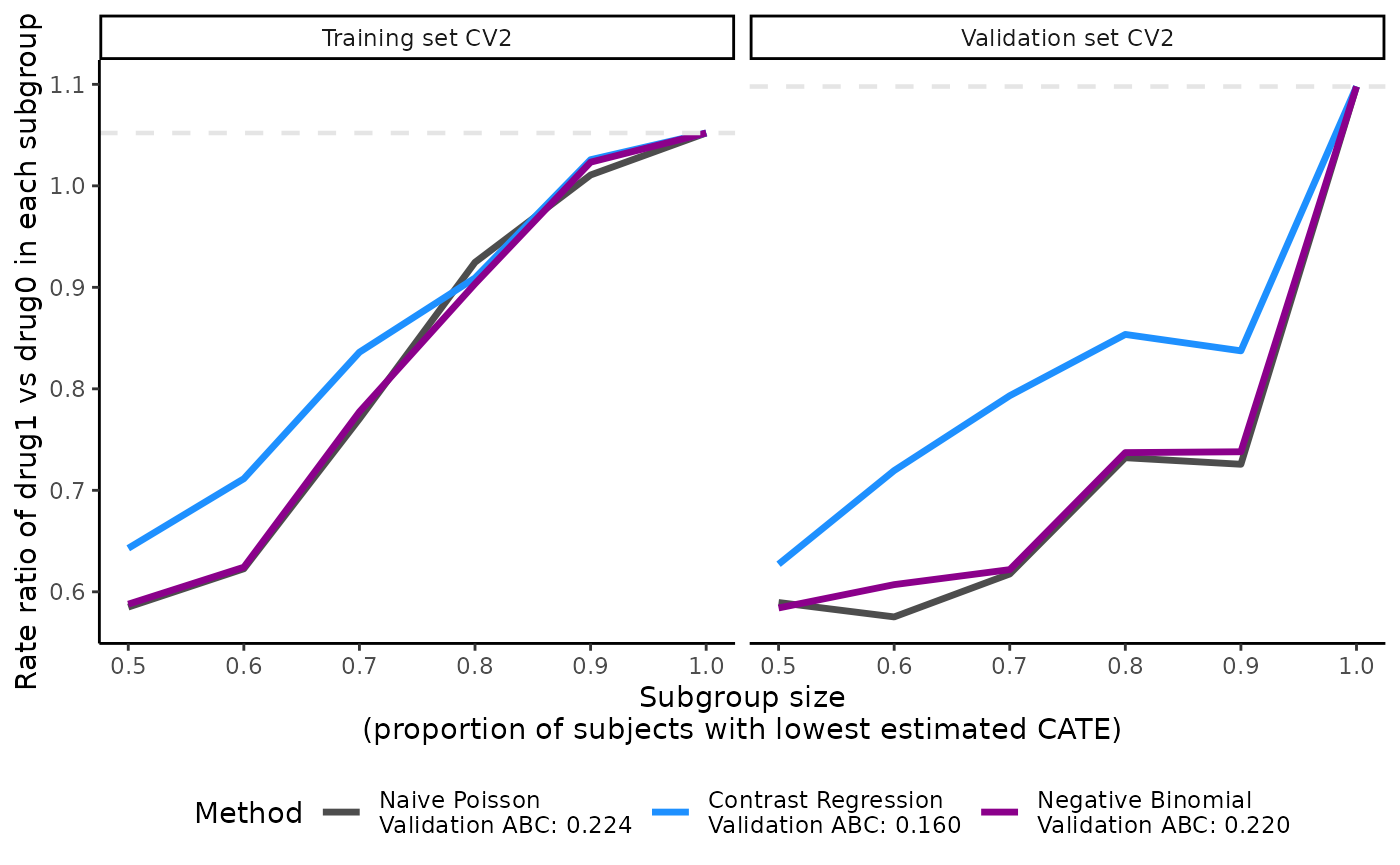
Same as abc(), the user can also choose to use the
median (instead of mean [default]) of the ATEs across CV iterations by
specifying the argument combine = “median” in
plot().
Visualization of the ATE in subgroups with
boxplot()
The ATEs of multi-category subgroups that are mutually exclusive can
be visualized as box plots, with one box plot for each scoring method.
Only validation results are visualized here. The x-axis is determined by
prop.multi and the y-axis is the estimated ATEs in each
subgroup. We specify the ylab argument accordingly. The
subgroups correspond to each row of the
ate.est.valid.group.cv result in
output_catecv, so in this example the subgroups are patient
with the 33% lowest (0-33%), middle 33% (33-66%), and highest 33%
(66-100%) estimated CATE scores. The box plot shows the distribution of
the ATEs over all cv.n CV iterations, instead of a summary
statistics like mean or median in plot().
boxplot(x = output_catecv,
ylab = "Rate ratio of drug1 vs drug0 in each subgroup")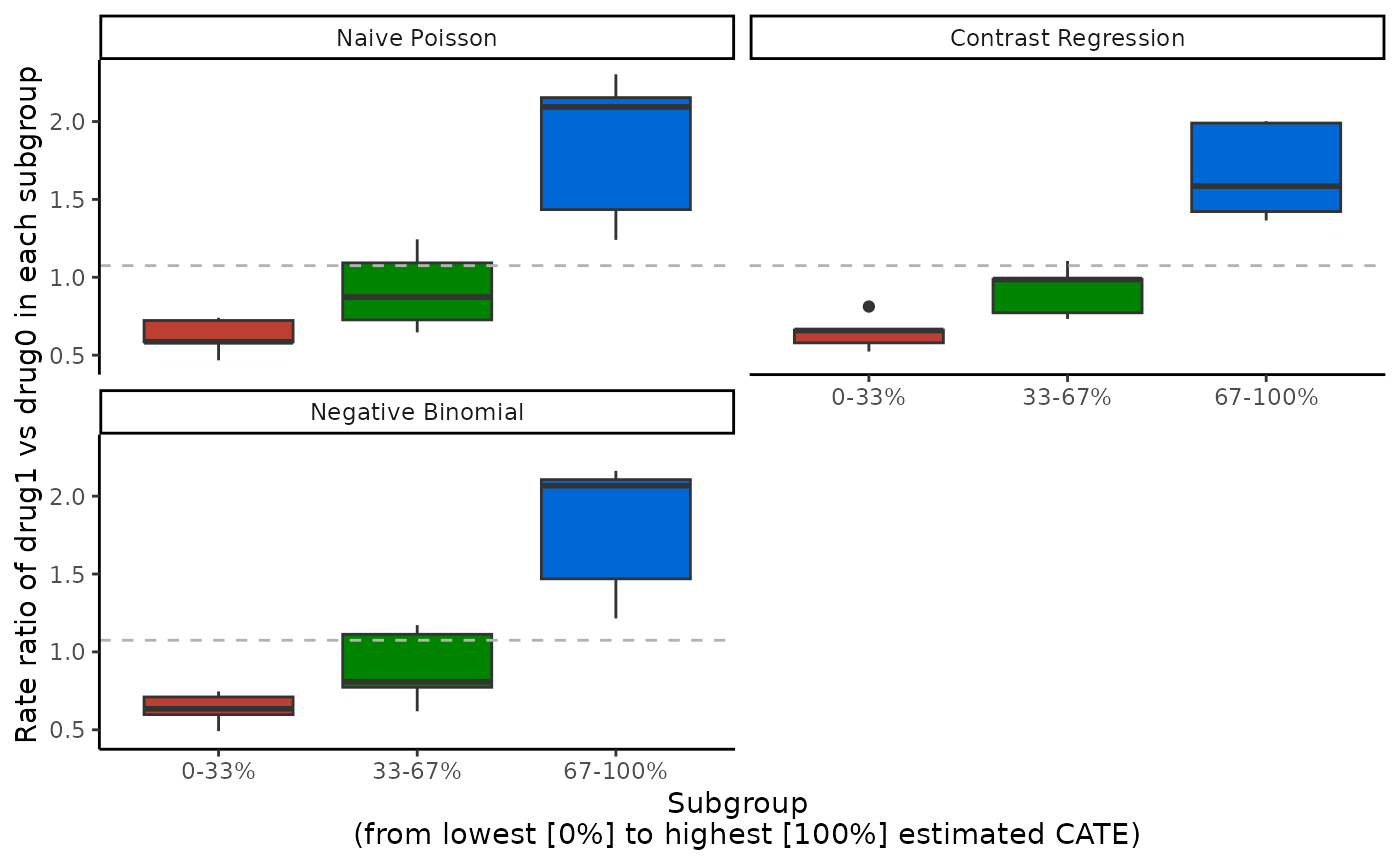
For this toy example, we can see why the two regressions method has the highest ABC and performs the best in the validation curves in the previous sections. Two regression has a decreasing RR as we go from the subgroup with the 33% lowest CATE scores (0-33%) to subgroup with the 33% highest CATE scores (66-100%), implying that there is some evidence of heterogeneous treatment effect and the CATE scores estimated with two regressions can distinguish the treatment heterogeneity in the data. In comparison, the other 3 methods seem to struggle with the validation data. Even tough they show different subgroups, we can see that the box plots correspond to the other 2 metrics. Note that the y-axis can have different scales for different scoring methods.
Although we provided 3 different metrics to summarize and visualize the
catecv()outputs, the user is encouraged to choose their own way of data wrangling that fits to their particular situation.
Other precmed vignettes in this series
1. Examples for count outcome
2. Examples for survival
outcome
3. Additional examples
4. Theoretical details