
Cross-validation of the conditional average treatment effect (CATE) score for count outcomes
Source:R/crossv.R
catecvcount.RdProvides doubly robust estimation of the average treatment effect (ATE) in
nested and mutually exclusive subgroups of patients defined by an estimated
conditional average treatment effect (CATE) score via cross-validation (CV).
The CATE score can be estimated with up to 5 methods among the following:
Poisson regression, boosting, two regressions, contrast regression, and
negative binomial (see score.method).
Usage
catecvcount(
data,
score.method,
cate.model,
ps.model,
ps.method = "glm",
initial.predictor.method = "boosting",
minPS = 0.01,
maxPS = 0.99,
higher.y = TRUE,
prop.cutoff = seq(0.5, 1, length = 6),
prop.multi = c(0, 1/3, 2/3, 1),
abc = TRUE,
train.prop = 3/4,
cv.n = 10,
error.max = 0.1,
max.iter = 5000,
xvar.smooth = NULL,
tree.depth = 2,
n.trees.boosting = 200,
B = 3,
Kfold = 5,
error.maxNR = 0.001,
max.iterNR = 150,
tune = c(0.5, 2),
seed = NULL,
plot.gbmperf = TRUE,
verbose = 0,
...
)Arguments
- data
A data frame containing the variables in the outcome and propensity score model; a data frame with
nrows (1 row per observation).- score.method
A vector of one or multiple methods to estimate the CATE score. Allowed values are:
'boosting','poisson','twoReg','contrastReg', and'negBin'.- cate.model
A formula describing the outcome model to be fitted. The outcome must appear on the left-hand side.
- ps.model
A formula describing the propensity score model to be fitted. The treatment must appear on the left-hand side. The treatment must be a numeric vector coded as 0 or 1. If data are from a randomized trial, specify
ps.modelas an intercept-only model.- ps.method
A character value for the method to estimate the propensity score. Allowed values include one of:
'glm'for logistic regression with main effects only (default), or'lasso'for a logistic regression with main effects and LASSO penalization on two-way interactions (added to the model if interactions are not specified inps.model). Relevant only whenps.modelhas more than one variable.- initial.predictor.method
A character vector for the method used to get initial outcome predictions conditional on the covariates in
cate.model. Only applies whenscore.methodincludes'twoReg'or'contrastReg'. Allowed values include one of'poisson'(fastest),'boosting'and'gam'. Default is'boosting'.- minPS
A numerical value between 0 and 1 below which estimated propensity scores should be truncated. Default is
0.01.- maxPS
A numerical value between 0 and 1 above which estimated propensity scores should be truncated. Must be strictly greater than
minPS. Default is0.99.- higher.y
A logical value indicating whether higher (
TRUE) or lower (FALSE) values of the outcome are more desirable. Default isTRUE.- prop.cutoff
A vector of numerical values between 0 and 1 specifying percentiles of the estimated log CATE scores to define nested subgroups. Each element represents the cutoff to separate observations in nested subgroups (below vs above cutoff). The length of
prop.cutoffis the number of nested subgroups. An equally-spaced sequence of proportions ending with 1 is recommended. Default isseq(0.5, 1, length = 6).- prop.multi
A vector of numerical values between 0 and 1 specifying percentiles of the estimated log CATE scores to define mutually exclusive subgroups. It should start with 0, end with 1, and be of
length(prop.multi) > 2. Each element represents the cutoff to separate the observations intolength(prop.multi) - 1mutually exclusive subgroups. Default isc(0, 1/3, 2/3, 1).- abc
A logical value indicating whether the area between curves (ABC) should be calculated at each cross-validation iterations, for each
score.method. Default isTRUE.- train.prop
A numerical value between 0 and 1 indicating the proportion of total data used for training. Default is
3/4.- cv.n
A positive integer value indicating the number of cross-validation iterations. Default is
10.- error.max
A numerical value > 0 indicating the tolerance (maximum value of error) for the largest standardized absolute difference in the covariate distributions or in the doubly robust estimated rate ratios between the training and validation sets. This is used to define a balanced training-validation splitting. Default is
0.1.- max.iter
A positive integer value indicating the maximum number of iterations when searching for a balanced training-validation split. Default is
5,000.- xvar.smooth
A vector of characters indicating the name of the variables used as the smooth terms if
initial.predictor.method = 'gam'. The variables must be selected from the variables listed incate.model. Default isNULL, which uses all variables incate.model.- tree.depth
A positive integer specifying the depth of individual trees in boosting (usually 2-3). Used only if
score.method = 'boosting'or ifinitial.predictor.method = 'boosting'withscore.method = 'twoReg'or'contrastReg'. Default is 2.- n.trees.boosting
A positive integer specifying the maximum number of trees in boosting (usually 100-1000). Used if
score.method = 'boosting'or ifinitial.predictor.method = 'boosting'withscore.method = 'twoReg'or'contrastReg'. Default is200.- B
A positive integer specifying the number of time cross-fitting is repeated in
score.method = 'twoReg'and'contrastReg'. Default is3.- Kfold
A positive integer specifying the number of folds used in cross-fitting to partition the data in
score.method = 'twoReg'and'contrastReg'. Default is5.- error.maxNR
A numerical value > 0 indicating the minimum value of the mean absolute error in Newton Raphson algorithm. Used only if
score.method = 'contrastReg'. Default is0.001.- max.iterNR
A positive integer indicating the maximum number of iterations in the Newton Raphson algorithm. Used only if
score.method = 'contrastReg'. Default is150.- tune
A vector of 2 numerical values > 0 specifying tuning parameters for the Newton Raphson algorithm.
tune[1]is the step size,tune[2]specifies a quantity to be added to diagonal of the slope matrix to prevent singularity. Used only ifscore.method = 'contrastReg'. Default isc(0.5, 2).- seed
An optional integer specifying an initial randomization seed for reproducibility. Default is
NULL, corresponding to no seed.- plot.gbmperf
A logical value indicating whether to plot the performance measures in boosting. Used only if
score.method = 'boosting'or ifscore.method = 'twoReg'or'contrastReg'andinitial.predictor.method = 'boosting'. Default isTRUE.- verbose
An integer value indicating what kind of intermediate progress messages should be printed.
0means no outputs.1means only progress bar and run time.2means progress bar, run time, and all errors and warnings. Default is0.- ...
Additional arguments for
gbm()
Value
Returns a list containing the following components saved as a "precmed" object:
ate.poisson: A list of results output ifscore.methodincludes'poisson':ate.est.train.high.cv: A matrix of numerical values withlength(prop.cutoff)rows andcv.ncolumns. The ith row/jth column cell contains the estimated ATE in the nested subgroup of high responders defined by CATE score above (ifhigher.y = TRUE) or below (ifhigher.y = FALSE) theprop.cutoff[i]x100% percentile of the estimated CATE score in the training set in the jth cross-validation iteration.ate.est.train.low.cv: A matrix of numerical values withlength(prop.cutoff) - 1rows andcv.ncolumns. The ith row/jth column cell contains the estimated ATE in the nested subgroup of low responders defined by CATE score below (ifhigher.y = TRUE) or above (ifhigher.y = FALSE) theprop.cutoff[i]x100% percentile of the estimated CATE score in the training set in the jth cross-validation iteration.ate.est.valid.high.cv: Same asate.est.train.high.cv, but in the validation set.ate.est.valid.low.cv: Same asate.est.train.low.cv, but in the validation set.ate.est.train.group.cv: A matrix of numerical values withlength(prop.multi) - 1rows andcv.ncolumns. The jth column contains the estimated ATE inlength(prop.multi) - 1mutually exclusive subgroups defined byprop.multiin the training set in jth cross-validation iteration.ate.est.valid.group.cv: Same asate.est.train.group.cv, but in the validation set.abc.valid: A vector of numerical values of lengthcv.n. The ith element returns the ABC of the validation curve in the ith cross-validation iteration. Only returned ifabc = TRUE.
ate.boosting: A list of results similar toate.poissonoutput ifscore.methodincludes'boosting'.ate.twoReg: A list of results similar toate.poissonoutput ifscore.methodincludes'twoReg'.ate.contrastReg: A list of results similar toate.poissonoutput ifscore.methodincludes'contrastReg'. This method has an additional element in the list of results:converge.contrastReg.cv: A vector of logical value of lengthcv.n. The ith element indicates whether the algorithm converged in the ith cross-validation iteration.
ate.negBin: A list of results similar toate.poissonoutput ifscore.methodincludes'negBin'.props: A list of 3 elements:prop.onlyhigh: The original argumentprop.cutoff, reformatted as necessary.prop.bi: The original argumentprop.cutoff, similar toprop.onlyhighbut reformatted to exclude 1.prop.multi: The original argumentprop.multi, reformatted as necessary to include 0 and 1.
overall.ate.valid: A vector of numerical values of lengthcv.n. The ith element contains the ATE in the validation set of the ith cross-validation iteration, estimated with the doubly robust estimator.overall.ate.train: A vector of numerical values of lengthcv.n. The ith element contains the ATE in the training set of the ith cross-validation iteration, estimated with the doubly robust estimator.fgam: The formula used in GAM ifinitial.predictor.method = 'gam'.higher.y: The originalhigher.yargument.abc: The originalabcargument.cv.n: The originalcv.nargument.response: The type of response. Always 'count' for this function.formulas:A list of 3 elements: (1)cate.modelargument, (2)ps.modelargument and (3) original labels of the left-hand side variable inps.model(treatment) if it was not 0/1.
Details
The CATE score represents an individual-level treatment effect expressed as a rate ratio for count outcomes. It can be estimated with boosting, Poisson regression, negative binomial regression, and the doubly robust estimator two regressions (Yadlowsky, 2020) applied separately by treatment group or with the other doubly robust estimator contrast regression (Yadlowsky, 2020) applied to the entire data set.
Internal CV is applied to reduce optimism in choosing the CATE estimation method that
captures the most treatment effect heterogeneity. The CV is applied by repeating the
following steps cv.n times:
Split the data into a training and validation set according to
train.prop. The training and validation sets must be balanced with respect to covariate distributions and doubly robust rate ratio estimates (seeerror.max).Estimate the CATE score in the training set with the specified scoring method.
Predict the CATE score in the validation set using the scoring model fitted from the training set.
Build nested subgroups of treatment responders in the training and validation sets, separately, and estimate the ATE within each nested subgroup. For each element i of
prop.cutoff(e.g.,prop.cutoff[i]= 0.6), take the following steps:Identify high responders as observations with the 60% (i.e.,
prop.cutoff[i]x100%) highest (ifhigher.y = TRUE) or lowest (ifhigher.y = FALSE) estimated CATE scores.Estimate the ATE in the subgroup of high responders using a doubly robust estimator.
Conversely, identify low responders as observations with the 40% (i.e., 1 -
prop.cutoff[i]x100%) lowest (ifhigher.y= TRUE) or highest (ifhigher.y= FALSE) estimated CATE scores.Estimate the ATE in the subgroup of low responders using a doubly robust estimator.
If
abc= TRUE, calculate the area between the ATE and the series of ATEs in nested subgroups of high responders in the validation set.Build mutually exclusive subgroups of treatment responders in the training and validation sets, separately, and estimate the ATE within each subgroup. Mutually exclusive subgroups are built by splitting the estimated CATE scores according to
prop.multi.
References
Yadlowsky, S., Pellegrini, F., Lionetto, F., Braune, S., & Tian, L. (2020). Estimation and validation of ratio-based conditional average treatment effects using observational data. Journal of the American Statistical Association, 1-18.. DOI: 10.1080/01621459.2020.1772080.
See also
plot.precmed(), boxplot.precmed(),
abc() methods for "precmed" objects,
and catefitcount() function.
Examples
# \donttest{
catecv <- catecvcount(data = countExample,
score.method = "poisson",
cate.model = y ~ age + female + previous_treatment +
previous_cost + previous_number_relapses +
offset(log(years)),
ps.model = trt ~ age + previous_treatment,
higher.y = FALSE,
cv.n = 5,
seed = 999,
plot.gbmperf = FALSE,
verbose = 1)
#> Warning: Variable trt was recoded to 0/1 with drug0->0 and drug1->1.
#>
|
| | 0%
#> cv = 1
#> splitting the data..
#> training..
#> validating..
#>
#> cv = 2
#> splitting the data..
#> training..
#> validating..
#>
#> cv = 3
#> splitting the data..
#> training..
#> validating..
#>
#> cv = 4
#> splitting the data..
#> training..
#> validating..
#>
#> cv = 5
#> splitting the data..
#> training..
#> validating..
#>
#> Total runtime : 11.32 secs
plot(catecv, ylab = "Rate ratio of drug1 vs drug0 in each subgroup")
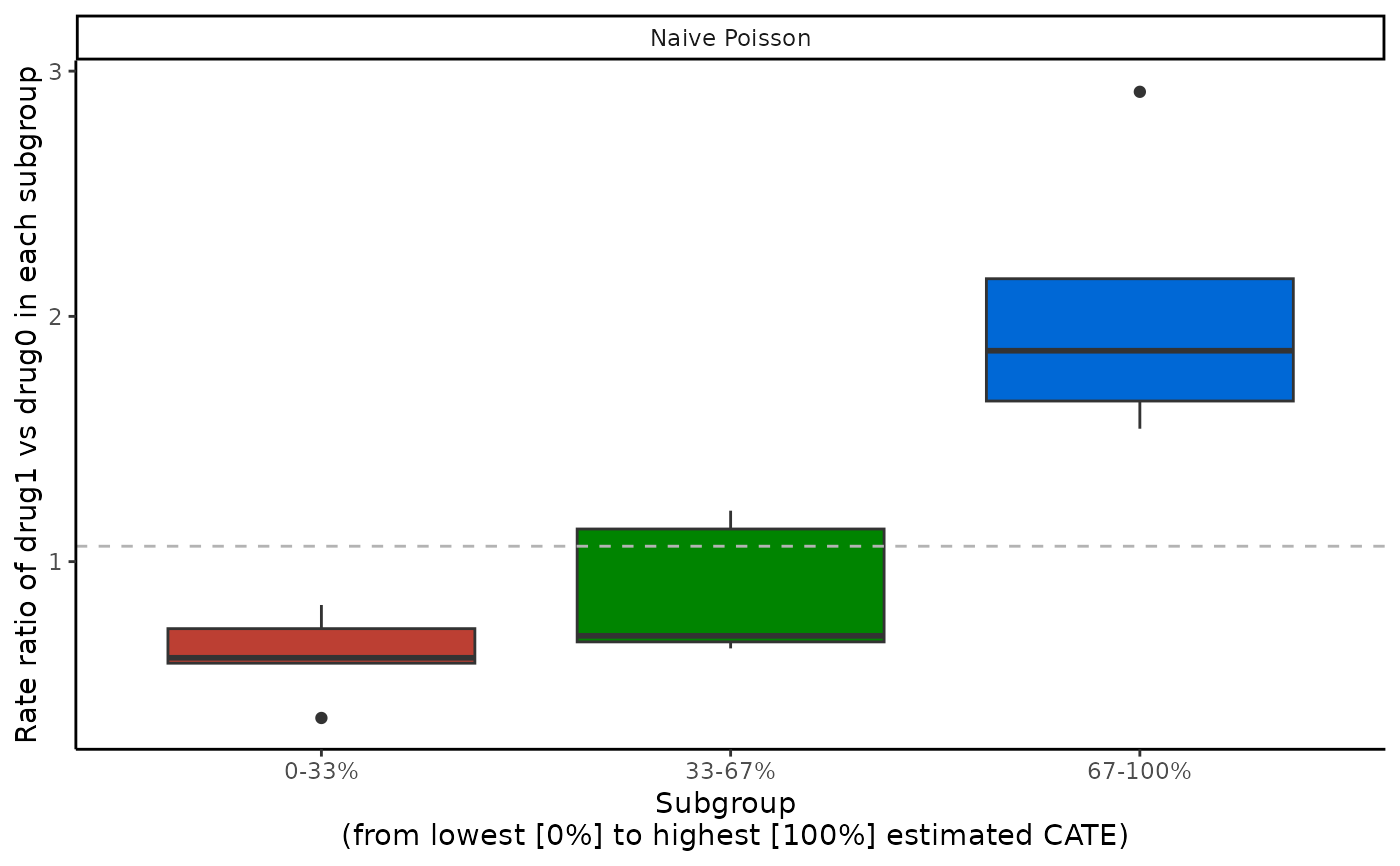
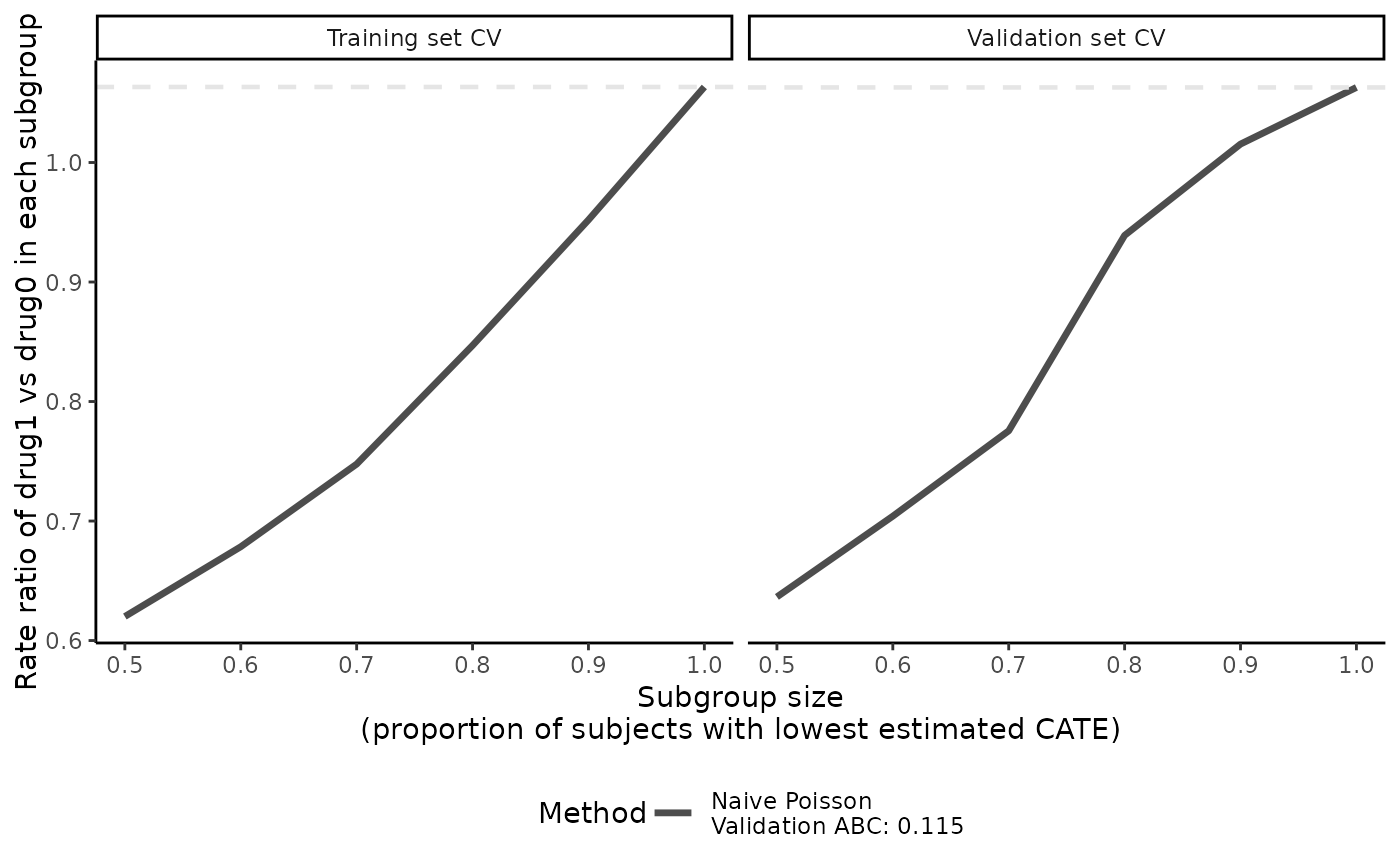 boxplot(catecv, ylab = "Rate ratio of drug1 vs drug0 in each subgroup")
boxplot(catecv, ylab = "Rate ratio of drug1 vs drug0 in each subgroup")
 abc(catecv)
#> cv1 cv2 cv3 cv4 cv5
#> poisson 0.1375899 0.1172791 0.1120313 0.06136285 0.1877103
# }
abc(catecv)
#> cv1 cv2 cv3 cv4 cv5
#> poisson 0.1375899 0.1172791 0.1120313 0.06136285 0.1877103
# }