source("resources/chapter 12/sim.r")
source("resources/chapter 12/fig_functions.r")
source("resources/chapter 12/mlmi.r")8 Dealing with irregular and informative visits
8.1 Introduction
We first load the relevant R scripts:
8.2 Example dataset
Below, we generate an example dataset that contains information on the treatment allocation x and three baseline covariates age, sex and edss (EDSS at treatment start). The discrete outcome y represents the Expanded Disability Status Scale (EDSS) score after time months of treatment exposure. Briefly, the EDSS is a semi-continuous measure that varies from 0 (no disability) to 10 (death).
set.seed(9843626)
dataset <- sim_data_EDSS(npatients = 500,
ncenters = 10,
follow_up = 12*5,
sd_a_t = 0.5,
baseline_EDSS = 1.3295,
sd_alpha_ij = 1.46,
sd_beta1_j = 0.20,
mean_age = 42.41,
sd_age = 10.53,
min_age = 18,
beta_age = 0.05,
beta_t = 0.014,
beta_t2 = 0,
delta_xt = 0,
delta_xt2 = 0,
p_female = 0.75,
beta_female = -0.2 ,
delta_xf = 0,
rho = 0.8,
corFUN = corAR1,
tx_alloc_FUN = treatment_alloc_confounding_v2)`summarise()` has grouped output by 'time'. You can override using the
`.groups` argument.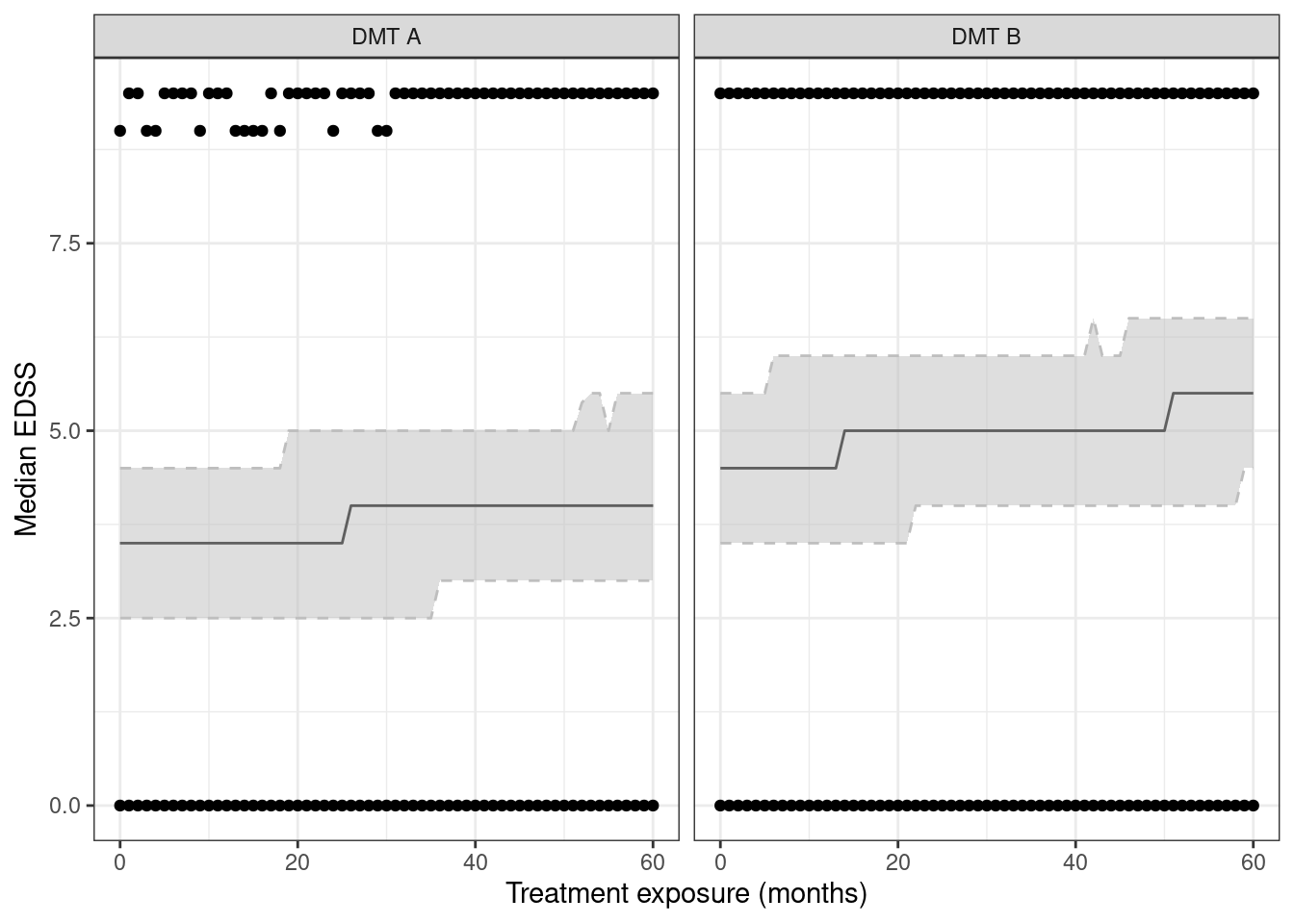
We remove the outcome y according to the informative visit process that depends on the received treatment, gender, and age.
dataset_visit <- censor_visits_a5(dataset, seed = 12345) %>%
dplyr::select(-y) %>%
mutate(time_x = time*x)In the censored data, a total of 17 out of 5000 patients have a visit at time=60.
8.3 Estimation of treatment effect
We will estimate the marginal treatment effect at time time=60.
8.3.1 Original data
origdat60 <- dataset %>% filter(time == 60)
# Predict probability of treatment allocation
fitps <- glm(x ~ age + sex + edss, family = 'binomial',
data = origdat60)
# Derive the propensity score
origdat60 <- origdat60 %>%
mutate(ipt = ifelse(x == 1, 1/predict(fitps, type = 'response'),
1/(1 - predict(fitps, type = 'response'))))
# Estimate
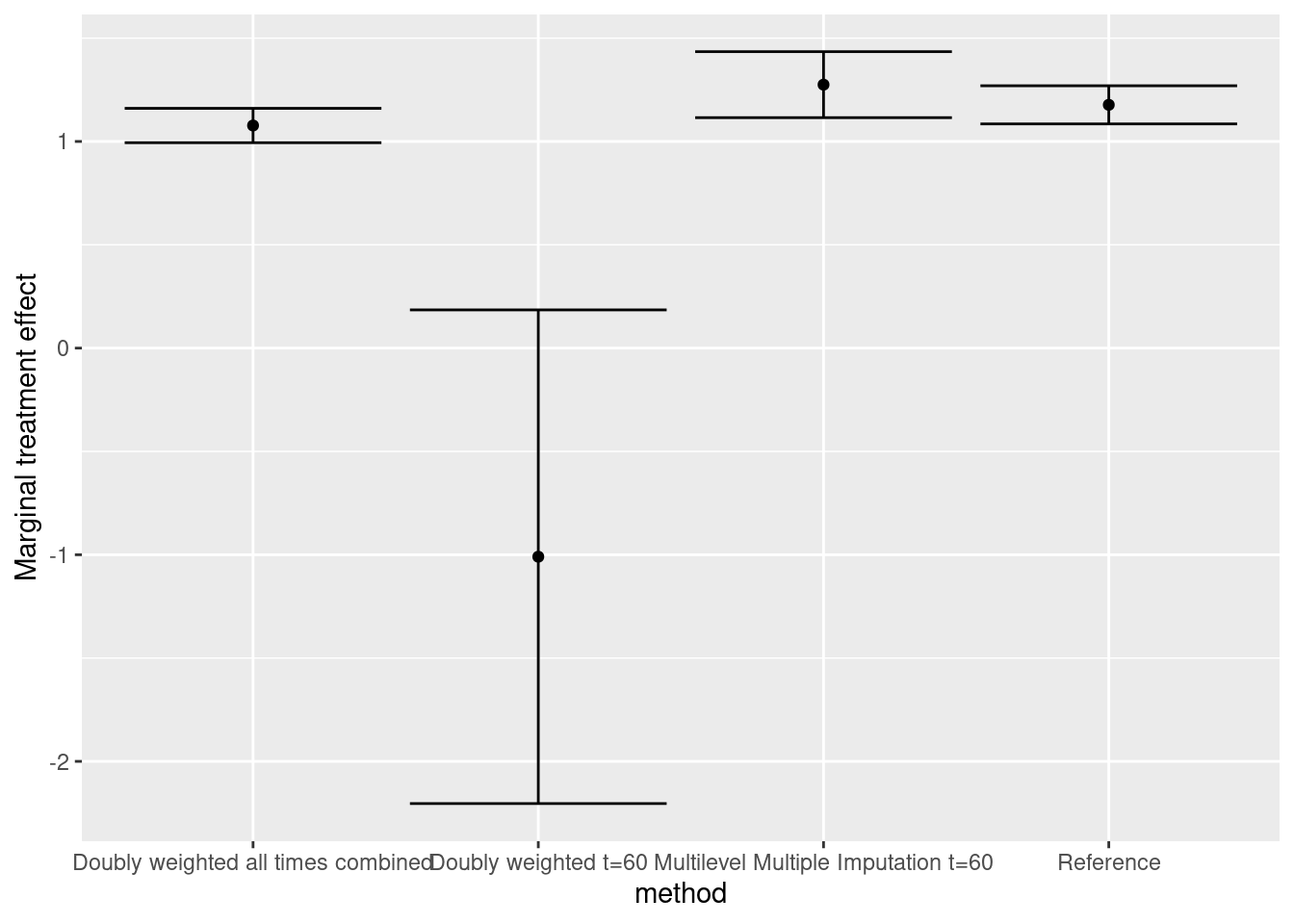
fit_ref_m <- tidy(lm(y ~ x, weight = ipt, data = origdat60), conf.int = TRUE) 8.3.2 Doubly-weighted marginal treatment effect
We here implement inverse probability of response weights into the estimating equations to adjust for nonrandom missingness Coulombe, Moodie, and Platt (2020).
obsdat60 <- dataset_visit %>%
mutate(visit = ifelse(is.na(y_obs),0,1)) %>%
filter(time == 60)
gamma <- glm(visit ~ x + sex + age + edss,
family = 'binomial', data = obsdat60)$coef
obsdat60 <- obsdat60 %>% mutate(rho_i = 1/exp(gamma["(Intercept)"] +
gamma["x"]*x +
gamma["sex"]*sex +
gamma["age"]*age))
# Predict probability of treatment allocation
fitps <- glm(x ~ age + sex + edss, family='binomial', data = obsdat60)
# Derive the propensity score
obsdat60 <- obsdat60 %>%
mutate(ipt = ifelse(x==1, 1/predict(fitps, type='response'),
1/(1 - predict(fitps, type='response'))))
fit_w <- tidy(lm(y_obs ~ x, weights = ipt*rho_i, data = obsdat60),
conf.int = TRUE)8.3.3 Multilevel multiple imputation
We adopt the imputation approach proposed by Debray et al. (2023). Briefly, we impute the entire vector of y_obs for all 61 potential visits and generate 10 imputed datasets. Note: mlmi currently does not support imputation of treatment-covariate interaction terms.
imp <- impute_y_mice_3l(dataset_visit, seed = 12345)We can now estimate the treatment effect in each imputed dataset
# Predict probability of treatment allocation
fitps <- glm(x ~ age + sex + edss, family = 'binomial', data = dataset_visit)
# Derive the propensity score
dataset_visit <- dataset_visit %>%
mutate(ipt = ifelse(x == 1, 1/predict(fitps, type = 'response'),
1/(1 - predict(fitps, type = 'response'))))
Q <- U <- rep(NA, 10) # Error variances
for (i in seq(10)) {
dati <- cbind(dataset_visit[,c("x","ipt","time")], y_imp = imp[,i]) %>%
filter(time == 60)
# Estimate
fit <- tidy(lm(y_imp ~ x, weight = ipt, data = dati), conf.int = TRUE)
Q[i] <- fit %>% filter(term == "x") %>% pull(estimate)
U[i] <- (fit %>% filter(term == "x") %>% pull(std.error))**2
}
fit_mlmi <- pool.scalar(Q = Q, U = U)8.4 Reproduce the results using all data to compute the marginal effect with IIV-weighted
8.4.1 Doubly -weighted marginal treatment effect total
obsdatall <- dataset_visit %>% mutate(visit = ifelse(is.na(y_obs),0,1))
gamma <- glm(visit ~ x + sex + age + edss, family = 'binomial', data = obsdatall)$coef
obsdatall <- obsdatall %>% mutate(rho_i = 1/exp(gamma["(Intercept)"] +
gamma["x"]*x +
gamma["sex"]*sex +
gamma["age"]*age))
# Predict probability of treatment allocation
fitps <- glm(x ~ age + sex + edss, family='binomial', data = obsdatall)
# Derive the propensity score
obsdatall <- obsdatall %>% mutate(ipt = ifelse(x==1, 1/predict(fitps, type='response'),
1/(1-predict(fitps, type='response'))))
fit_w <- tidy(lm(y_obs ~ x, weights = ipt*rho_i, data = obsdatall), conf.int = TRUE)8.5 Results
Version info
This chapter was rendered using the following version of R and its packages:
R version 4.2.3 (2023-03-15)
Platform: x86_64-pc-linux-gnu (64-bit)
Running under: Ubuntu 22.04.4 LTS
Matrix products: default
BLAS: /usr/lib/x86_64-linux-gnu/openblas-pthread/libblas.so.3
LAPACK: /usr/lib/x86_64-linux-gnu/openblas-pthread/libopenblasp-r0.3.20.so
locale:
[1] LC_CTYPE=C.UTF-8 LC_NUMERIC=C LC_TIME=C.UTF-8
[4] LC_COLLATE=C.UTF-8 LC_MONETARY=C.UTF-8 LC_MESSAGES=C.UTF-8
[7] LC_PAPER=C.UTF-8 LC_NAME=C LC_ADDRESS=C
[10] LC_TELEPHONE=C LC_MEASUREMENT=C.UTF-8 LC_IDENTIFICATION=C
attached base packages:
[1] stats graphics grDevices utils datasets methods base
other attached packages:
[1] sparseMVN_0.2.2 truncnorm_1.0-9 MASS_7.3-58.2 nlme_3.1-162
[5] mice_3.16.0 ggplot2_3.5.0 broom_1.0.5 dplyr_1.1.4
loaded via a namespace (and not attached):
[1] shape_1.4.6.1 tidyselect_1.2.1 xfun_0.42 purrr_1.0.2
[5] splines_4.2.3 lattice_0.20-45 colorspace_2.1-0 vctrs_0.6.5
[9] generics_0.1.3 htmltools_0.5.7 yaml_2.3.8 pan_1.9
[13] utf8_1.2.4 survival_3.5-3 rlang_1.1.3 jomo_2.7-6
[17] pillar_1.9.0 nloptr_2.0.3 withr_3.0.0 glue_1.7.0
[21] foreach_1.5.2 lifecycle_1.0.4 munsell_0.5.0 gtable_0.3.4
[25] htmlwidgets_1.6.4 codetools_0.2-19 evaluate_0.23 labeling_0.4.3
[29] knitr_1.45 fastmap_1.1.1 fansi_1.0.6 Rcpp_1.0.12
[33] scales_1.3.0 backports_1.4.1 jsonlite_1.8.8 farver_2.1.1
[37] lme4_1.1-35.1 digest_0.6.35 grid_4.2.3 cli_3.6.2
[41] tools_4.2.3 magrittr_2.0.3 glmnet_4.1-8 tibble_3.2.1
[45] tidyr_1.3.1 pkgconfig_2.0.3 ellipsis_0.3.2 Matrix_1.6-5
[49] minqa_1.2.6 rmarkdown_2.26 iterators_1.0.14 mitml_0.4-5
[53] R6_2.5.1 boot_1.3-28.1 rpart_4.1.19 nnet_7.3-18
[57] compiler_4.2.3