library(dplyr)
library(gemtc)
library(netmeta)6 Systematic review and meta-analysis of Real-World Evidence
6.1 Introduction
We first load the required packages
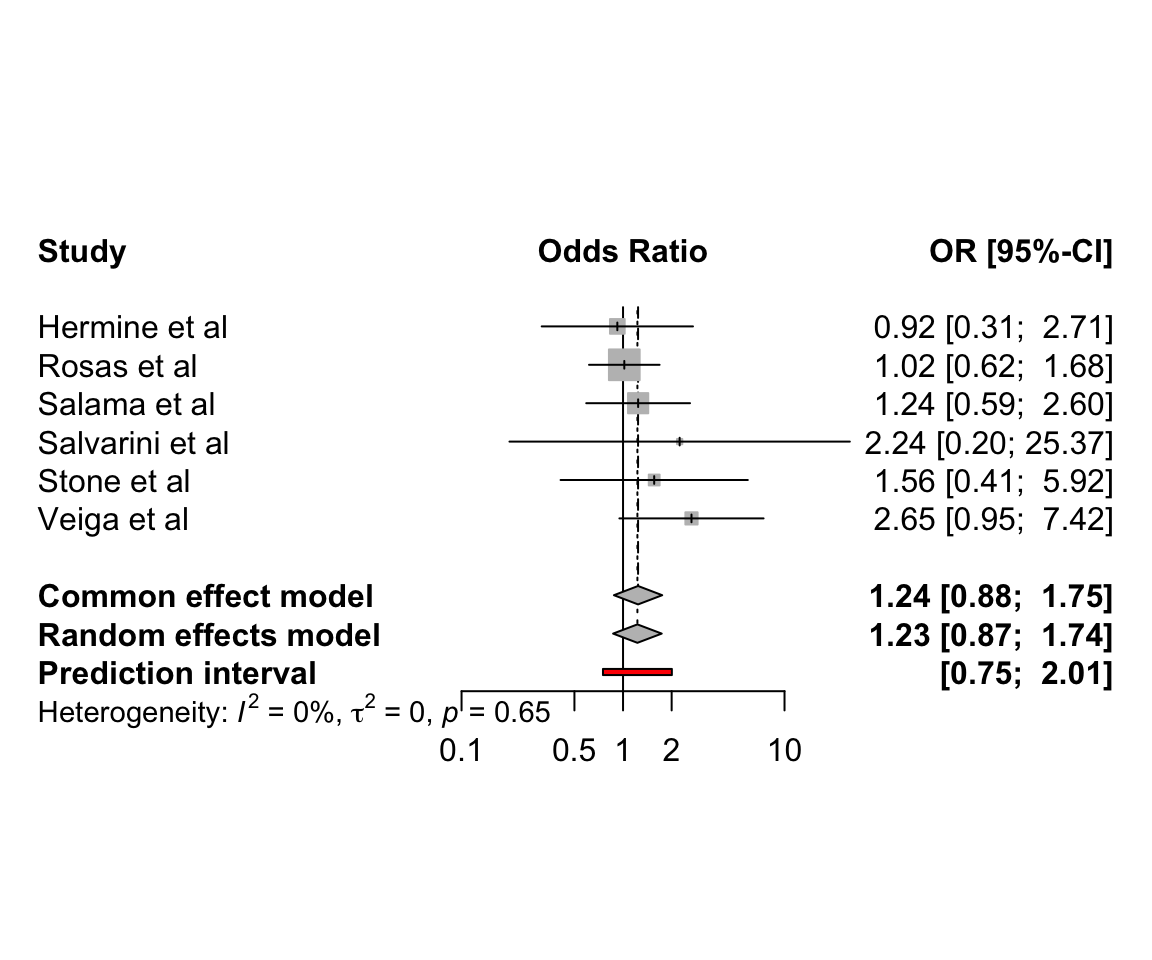
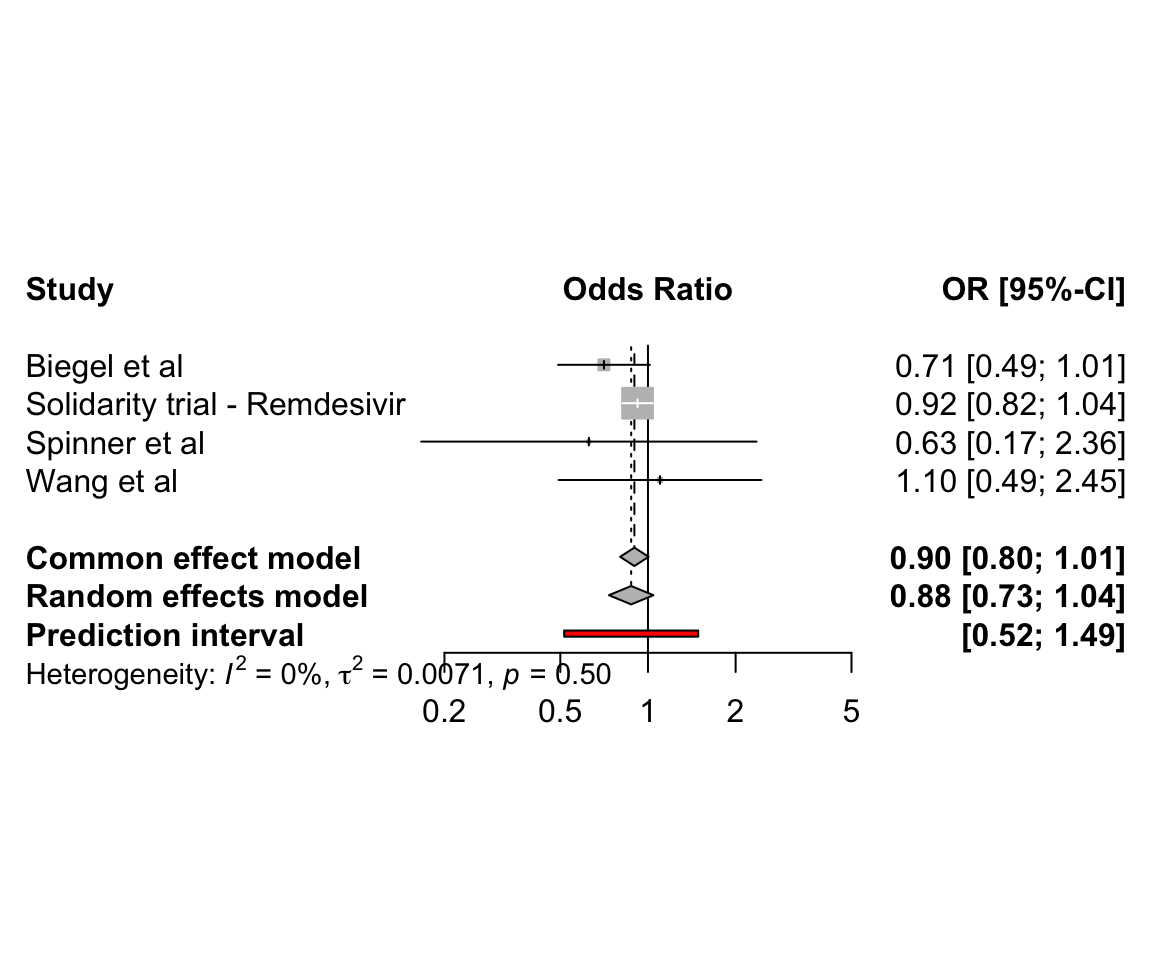
6.2 Pairwise meta-analysis of clinical trials
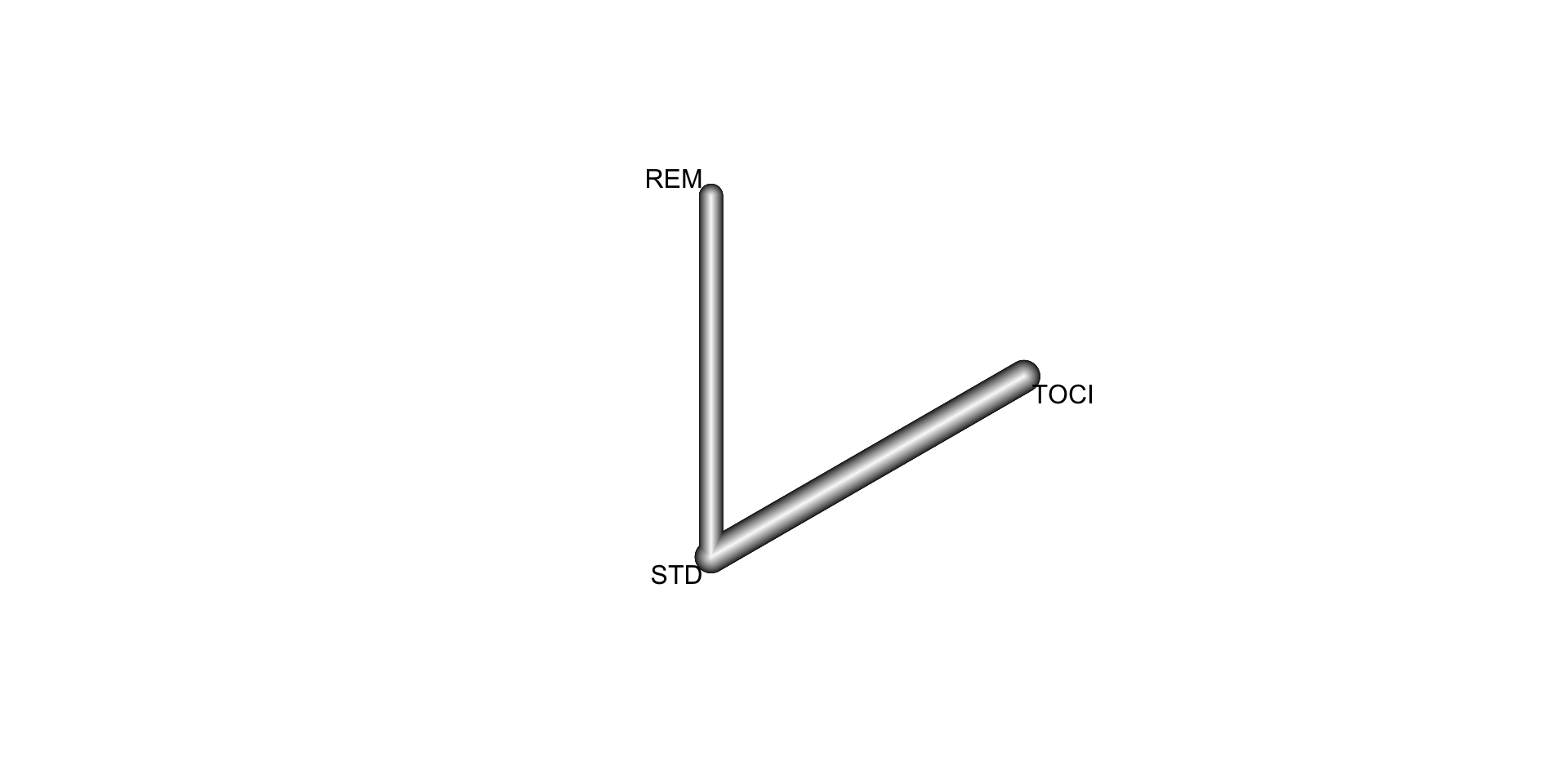
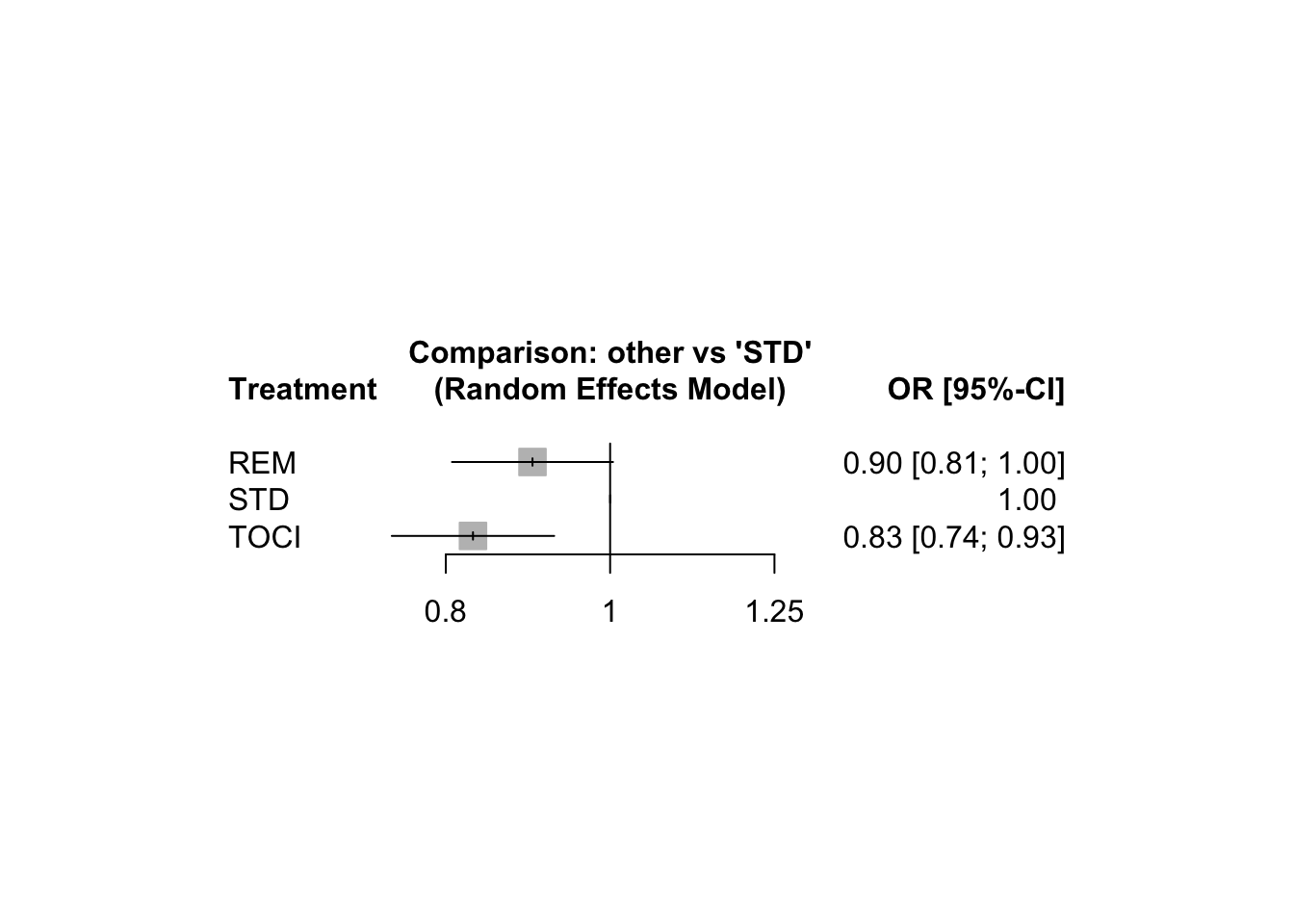
6.3 Network meta-analysis of clinical trials
We here use the R packages netmeta for conducting a frequentist network meta-analysis. A detailed tutorial on the use of netmeta is available from the book Doing Meta-Analysis with R: A Hands-On Guide.
6.3.2 Pharmacologic treatments for chronic obstructive pulmonary disease
In this example, we consider the resuls from a systematic review of randomized controlled trials on pharmacologic treatments for chronic obstructive pulmonary disease (Baker, Baker, and Coleman 2009). The primary outcome, occurrence of one or more episodes of COPD exacerbation, is binary (yes / no). For this outcome, five drug treatments (fluticasone, budesonide, salmeterol, formoterol, tiotropium) and two combinations (fluticasone + salmeterol, budesonide + formoterol) were compared to placebo. The authors considered the two combinations as separate treatments instead of evaluating the individual components.
data(Baker2009)| study | year | id | treatment | exac | total |
|---|---|---|---|---|---|
| Llewellyn-Jones 1996 | 1996 | 1 | Fluticasone | 0 | 8 |
| Llewellyn-Jones 1996 | 1996 | 1 | Placebo | 3 | 8 |
| Boyd 1997 | 1997 | 2 | Salmeterol | 47 | 229 |
| Boyd 1997 | 1997 | 2 | Placebo | 59 | 227 |
| Paggiaro 1998 | 1998 | 3 | Fluticasone | 45 | 142 |
| Paggiaro 1998 | 1998 | 3 | Placebo | 51 | 139 |
Baker <- pairwise(treat = treatment,
event = exac,
n = total,
studlab = id,
sm = "OR",
data = Baker2009)
NMA.COPD <- netmeta(TE = TE, seTE = seTE, treat1 = treat1, treat2 = treat2,
studlab = studlab, data = Baker, sm = "OR", ref = "Placebo",
comb.random = TRUE)Warning: Comparisons with missing TE / seTE or zero seTE not considered in
network meta-analysis.Comparisons not considered in network meta-analysis:
studlab treat1 treat2 TE seTE
39 Fluticasone+Salmeterol Placebo NA NA
39 Fluticasone+Salmeterol Salmeterol NA NA
39 Salmeterol Placebo NA NAnetgraph(NMA.COPD)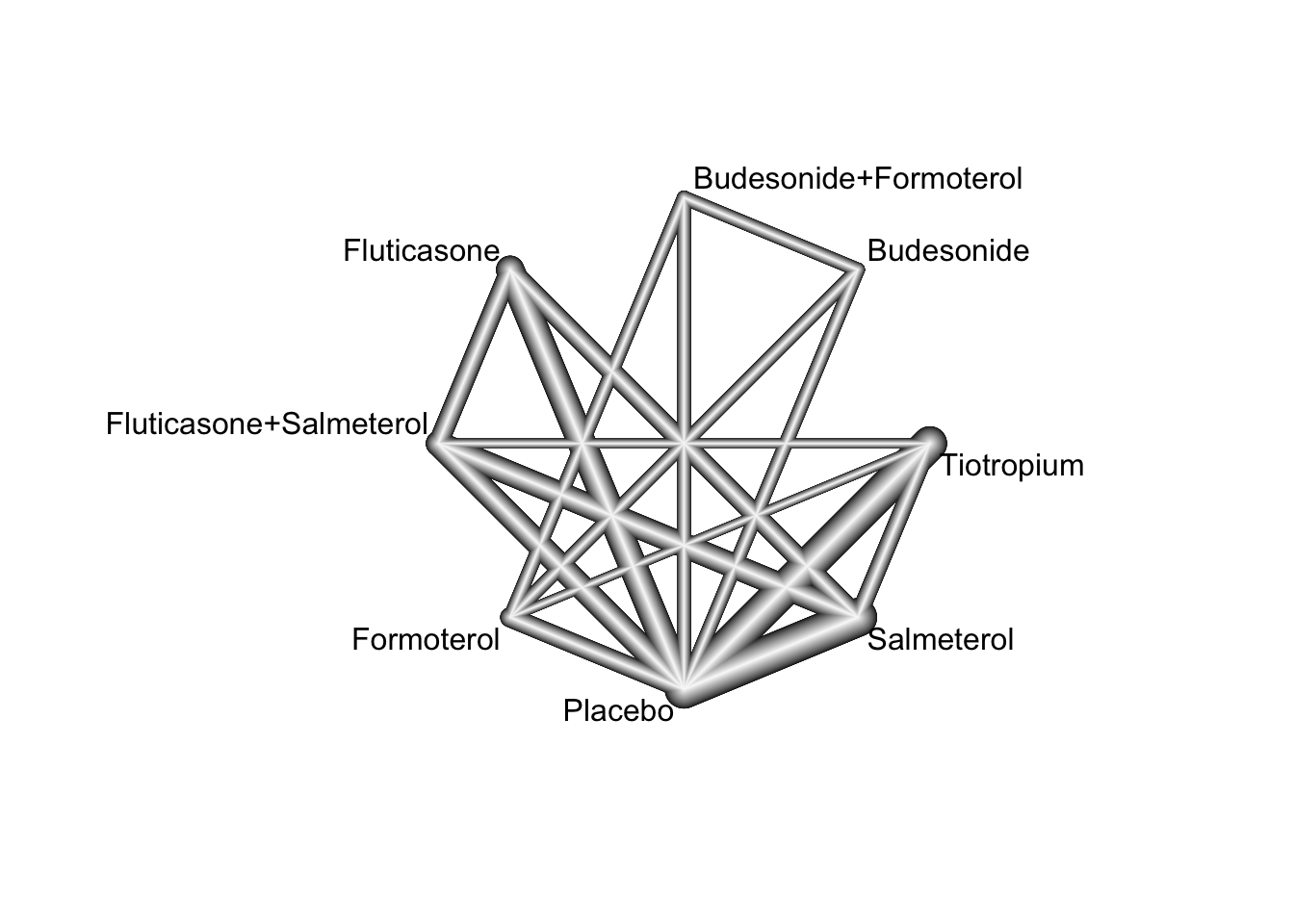
We can test for local network inconsistency as follows:
netsplit(NMA.COPD)By default, indirect estimates are derived using the back-calculation method. A comparison of direct and indirect treatment effect estimates (depicted as odds ratio) from the random effects network meta-analysis model is given in the table below:
| comparison | k | nma | direct | indirect | RoR | p.RoR |
|---|---|---|---|---|---|---|
| Budesonide:Budesonide+Formoterol | 2 | 1.20 | 1.20 | 745.35 | 0.00 | 0.28 |
| Budesonide:Fluticasone | 0 | 0.93 | . | 0.93 | . | . |
| Budesonide:Fluticasone+Salmeterol | 0 | 1.07 | . | 1.07 | . | . |
| Budesonide:Formoterol | 2 | 0.80 | 0.74 | 1.39 | 0.53 | 0.18 |
| Budesonide:Placebo | 2 | 0.80 | 0.85 | 0.52 | 1.63 | 0.33 |
| Budesonide:Salmeterol | 0 | 1.00 | . | 1.00 | . | . |
| Budesonide:Tiotropium | 0 | 1.17 | . | 1.17 | . | . |
| Budesonide+Formoterol:Fluticasone | 0 | 0.77 | . | 0.77 | . | . |
| Budesonide+Formoterol:Fluticasone+Salmeterol | 0 | 0.89 | . | 0.89 | . | . |
| Budesonide+Formoterol:Formoterol | 2 | 0.67 | 0.63 | 0.94 | 0.67 | 0.37 |
| Budesonide+Formoterol:Placebo | 2 | 0.67 | 0.72 | 0.39 | 1.83 | 0.21 |
| Budesonide+Formoterol:Salmeterol | 0 | 0.83 | . | 0.83 | . | . |
| Budesonide+Formoterol:Tiotropium | 0 | 0.97 | . | 0.97 | . | . |
| Fluticasone:Fluticasone+Salmeterol | 4 | 1.15 | 1.09 | 1.43 | 0.77 | 0.20 |
| Fluticasone:Formoterol | 0 | 0.86 | . | 0.86 | . | . |
| Fluticasone:Placebo | 8 | 0.86 | 0.90 | 0.67 | 1.34 | 0.15 |
| Fluticasone:Salmeterol | 4 | 1.08 | 1.07 | 1.09 | 0.98 | 0.90 |
| Fluticasone:Tiotropium | 0 | 1.26 | . | 1.26 | . | . |
| Fluticasone+Salmeterol:Formoterol | 0 | 0.75 | . | 0.75 | . | . |
| Fluticasone+Salmeterol:Placebo | 6 | 0.75 | 0.80 | 0.67 | 1.19 | 0.23 |
| Fluticasone+Salmeterol:Salmeterol | 6 | 0.94 | 0.91 | 1.01 | 0.90 | 0.53 |
| Fluticasone+Salmeterol:Tiotropium | 1 | 1.09 | 1.14 | 1.08 | 1.05 | 0.77 |
| Formoterol:Placebo | 4 | 1.00 | 0.96 | 2.79 | 0.34 | 0.09 |
| Formoterol:Salmeterol | 0 | 1.25 | . | 1.25 | . | . |
| Formoterol:Tiotropium | 1 | 1.46 | 4.20 | 1.39 | 3.03 | 0.07 |
| Salmeterol:Placebo | 15 | 0.80 | 0.79 | 0.85 | 0.93 | 0.65 |
| Tiotropium:Placebo | 12 | 0.69 | 0.70 | 0.63 | 1.11 | 0.50 |
| Salmeterol:Tiotropium | 3 | 1.17 | 1.14 | 1.18 | 0.96 | 0.80 |
6.3.3 Advanced Therapies for Ulcerative Colitis
In this example, we consider a systematic literature review of Phase 3 randomized controlled trials investigating the following advanced therapies: infliximab, adalimumab, vedolizumab, golimumab, tofacitinib, ustekinumab, filgotinib, ozanimod, and upadacitinib (Panaccione et al. 2023). This review included 48 RCTs, from which 23 were found eligible for inclusion in a network meta-analysis. The included RCT populations were largely comparable in their baseline characteristics, though some heterogeneity was noted in weight, disease duration, extent of disease, and concomitant medications. A risk of bias assessment showed a low risk of bias for all included RCTs, which were all industry sponsored.
We here focus on the synthesis of 18 trials that contributed efficacy data for induction in bio-naive populations. The following FDA- and/or EMA-approved biologic or SMD doses were investigated:
- Adalimumab subcutaneous 160 mg at week 0, 80 mg at week 2, and 40 mg at week 4 (
ADA160/80) - Infliximab intravenous 5 mg/kg (
INF5) at weeks 0, 2, and 6 then every 8 weeks - Infliximab intravenous 10 mg/kg (
INF10) at weeks 0, 2, and 6 then every 8 weeks - Filgotinib oral 100 mg once daily (
FIL100) - Filgotinib oral 200 mg once daily (
FIL200) - Golimumab subcutaneous 200 mg at week 0 and 100 mg at week 2 (
GOL200/100) - Ozanimod oral 0.23 mg once daily for 4 days, 0.46 mg once daily for 3 days, then 0.92 mg once daily (
OZA0.92) - Tofacitinib oral 10 mg twice daily for 8 weeks (
TOF10) - Upadacitinib oral 45 mg once daily for 8 weeks (
UPA45) - Ustekinumab intravenous 6 mg/kg at week 0 (
UST6) - Vedolizumab intravenous 300 mg at weeks 0, 2, and 6 (
VED300)
The reference treatment is placebo (PBO).
| studlab | treat1 | treat2 | event1 | n1 | event2 | n2 |
|---|---|---|---|---|---|---|
| ACT-1 | INF10 | INF5 | 39 | 122 | 47 | 121 |
| ACT-1 | INF10 | PBO | 39 | 122 | 18 | 121 |
| ACT-1 | INF5 | PBO | 47 | 121 | 18 | 121 |
| ACT-2 | INF10 | INF5 | 33 | 120 | 41 | 121 |
| ACT-2 | INF10 | PBO | 33 | 120 | 7 | 123 |
| ACT-2 | INF5 | PBO | 41 | 121 | 7 | 123 |
| GEMINI 1 | VED300 | PBO | 30 | 130 | 5 | 76 |
| Japic CTI-060298 | INF5 | PBO | 21 | 104 | 11 | 104 |
| Jiang 2015 | INF5 | PBO | 22 | 41 | 9 | 41 |
| M10-447 | ADA160/80 | PBO | 9 | 90 | 11 | 96 |
| NCT01551290 | INF5 | PBO | 11 | 50 | 5 | 49 |
| NCT02039505 | VED300 | PBO | 22 | 79 | 6 | 41 |
| OCTAVE 1 | TOF10 | PBO | 56 | 222 | 9 | 57 |
| OCTAVE 2 | TOF10 | PBO | 43 | 195 | 4 | 47 |
| PURSUIT-SC | GOL200/100 | PBO | 45 | 253 | 16 | 251 |
| SELECTION | FIL100 | FIL200 | 47 | 277 | 60 | 245 |
| SELECTION | FIL100 | PBO | 47 | 277 | 17 | 137 |
| SELECTION | FIL200 | PBO | 60 | 245 | 17 | 137 |
| TRUE NORTH | OZA0.92 | PBO | 66 | 299 | 10 | 151 |
| U-ACCOMPLISH | UPA45 | PBO | 54 | 166 | 3 | 81 |
| U-ACHIEVE Study 2 | UPA45 | PBO | 41 | 145 | 4 | 72 |
| ULTRA-1 | ADA160/80 | PBO | 24 | 130 | 12 | 130 |
| ULTRA-2 | ADA160/80 | PBO | 32 | 150 | 16 | 145 |
| UNIFI | UST6 | PBO | 27 | 147 | 15 | 151 |
The corresponding network is displayed below:
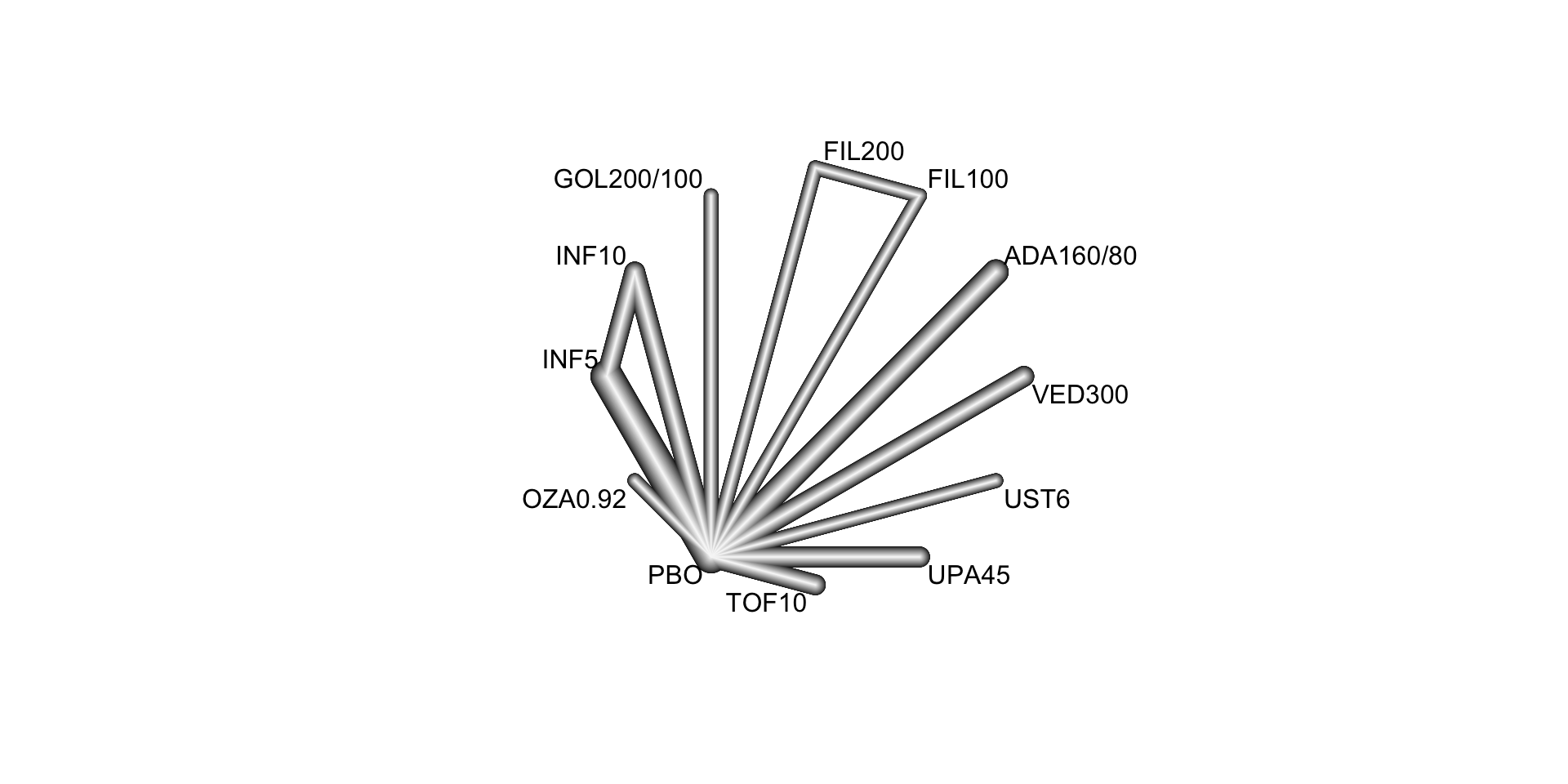
Below, we conduct a random effects network meta-analysis of the reported study effects (expressed as odds ratio) and consider placebo (treat = "PBO") as the control treatment.
NMA.uc <- netmeta(TE = TE, seTE = seTE, treat1 = treat1, treat2 = treat2,
studlab = studlab, data = UlcerativeColitis, sm = "OR",
ref = "PBO", common = FALSE, comb.random = TRUE)
NMA.ucAll treatments except FIL100 and UST6 are significantly more efficacious than PBO at inducing clinical remission. We can now estimate the probabilities of each treatment being at each possible rank and the SUCRAs (Surface Under the Cumulative RAnking curve):
sucra.uc <- rankogram(NMA.uc, nsim = 100, random = TRUE, common = FALSE,
small.values = "undesirable")
# Exctract the SUCRA values
sucra.uc$ranking.random ADA160/80 FIL100 FIL200 GOL200/100 INF10 INF5 OZA0.92
0.28090909 0.14363636 0.43272727 0.65454545 0.61727273 0.73727273 0.78909091
PBO TOF10 UPA45 UST6 VED300
0.01727273 0.37363636 0.98454545 0.37363636 0.59545455 These results indicate that 98.5% of the evaluated treatments are worse than UPA45.
Version info
This chapter was rendered using the following version of R and its packages:
R version 4.3.0 (2023-04-21)
Platform: aarch64-apple-darwin20 (64-bit)
Running under: macOS 14.2.1
Matrix products: default
BLAS: /Library/Frameworks/R.framework/Versions/4.3-arm64/Resources/lib/libRblas.0.dylib
LAPACK: /Library/Frameworks/R.framework/Versions/4.3-arm64/Resources/lib/libRlapack.dylib; LAPACK version 3.11.0
locale:
[1] en_US.UTF-8/en_US.UTF-8/en_US.UTF-8/C/en_US.UTF-8/en_US.UTF-8
time zone: Europe/Amsterdam
tzcode source: internal
attached base packages:
[1] stats graphics grDevices utils datasets methods base
other attached packages:
[1] netmeta_2.8-2 meta_6.5-0 gemtc_1.0-2 coda_0.19-4
[5] dplyr_1.1.4 kableExtra_1.3.4
loaded via a namespace (and not attached):
[1] magic_1.6-1 utf8_1.2.4 generics_0.1.3
[4] xml2_1.3.6 stringi_1.8.2 lattice_0.22-5
[7] lme4_1.1-35.1 digest_0.6.33 magrittr_2.0.3
[10] evaluate_0.23 grid_4.3.0 mvtnorm_1.2-4
[13] CompQuadForm_1.4.3 fastmap_1.1.1 plyr_1.8.9
[16] jsonlite_1.8.8 Matrix_1.6-4 httr_1.4.7
[19] rvest_1.0.3 fansi_1.0.6 viridisLite_0.4.2
[22] scales_1.3.0 codetools_0.2-19 numDeriv_2016.8-1.1
[25] abind_1.4-5 cli_3.6.1 rlang_1.1.2
[28] munsell_0.5.0 splines_4.3.0 withr_2.5.2
[31] yaml_2.3.7 tools_4.3.0 nloptr_2.0.3
[34] metafor_4.4-0 minqa_1.2.6 ggplot2_3.4.4
[37] colorspace_2.1-0 mathjaxr_1.6-0 webshot_0.5.5
[40] boot_1.3-28.1 vctrs_0.6.5 R6_2.5.1
[43] lifecycle_1.0.4 stringr_1.5.1 htmlwidgets_1.6.4
[46] MASS_7.3-60 pkgconfig_2.0.3 gtable_0.3.4
[49] pillar_1.9.0 glue_1.6.2 Rcpp_1.0.11
[52] systemfonts_1.0.5 highr_0.10 xfun_0.41
[55] tibble_3.2.1 tidyselect_1.2.0 rstudioapi_0.15.0
[58] knitr_1.45 htmltools_0.5.7 nlme_3.1-164
[61] igraph_1.5.1 rmarkdown_2.25 svglite_2.1.3
[64] compiler_4.3.0 metadat_1.2-0